| Deletions are marked like this. | Additions are marked like this. |
| Line 40: | Line 40: |
| -e number of streamlines to be used for the eigendecomposition of normalized cuts algorithm. The default is 500. Using less number of streamlines will fasten substantially the algorithm at the expense of accuracy. For a quick test 50 could be ok, but in practice less than 300 is not recommended. | -e number of streamlines to be used for the eigendecomposition of normalized cuts algorithm. The default is 500. Using less number of streamlines will fasten substantially the algorithm at the expense of accuracy. For a quick test 50 could be ok, but in practice, less than 300 is not recommended. |
| Line 46: | Line 46: |
| -e number of streamlines to be used for the eigendecomposition of normalized cuts algorithm. The default is 500. Using less number of streamlines will fasten substantially the algorithm at the expense of accuracy. For a quick test 50 could be ok, but in practice less than 300 is not recommended (See Siless et al, 2018). | -e number of streamlines to be used for the eigendecomposition of normalized cuts algorithm. The default is 500. Using less number of streamlines will fasten substantially the algorithm at the expense of accuracy. For a quick test 50 could be ok, but in practice, less than 300 is not recommended. |
| Line 48: | Line 48: |
|
-d directional neighbors to be used, the default is "a" all (26), diagonal "d" is 14 and straight "s" is 6. (See Siless et al 2018). |
-d directional neighbors to be used, the default is "a" all (26), diagonal "d" is 14 and straight "s" is 6. |
| Line 54: | Line 53: |
| You can find one-to-one correspondences using AnatomiCuts_correspondences. | You can find one-to-one correspondences using [[AnatomiCuts_correspondences| AnatomiCuts_correspondences]]. . |
AnatomiCuts
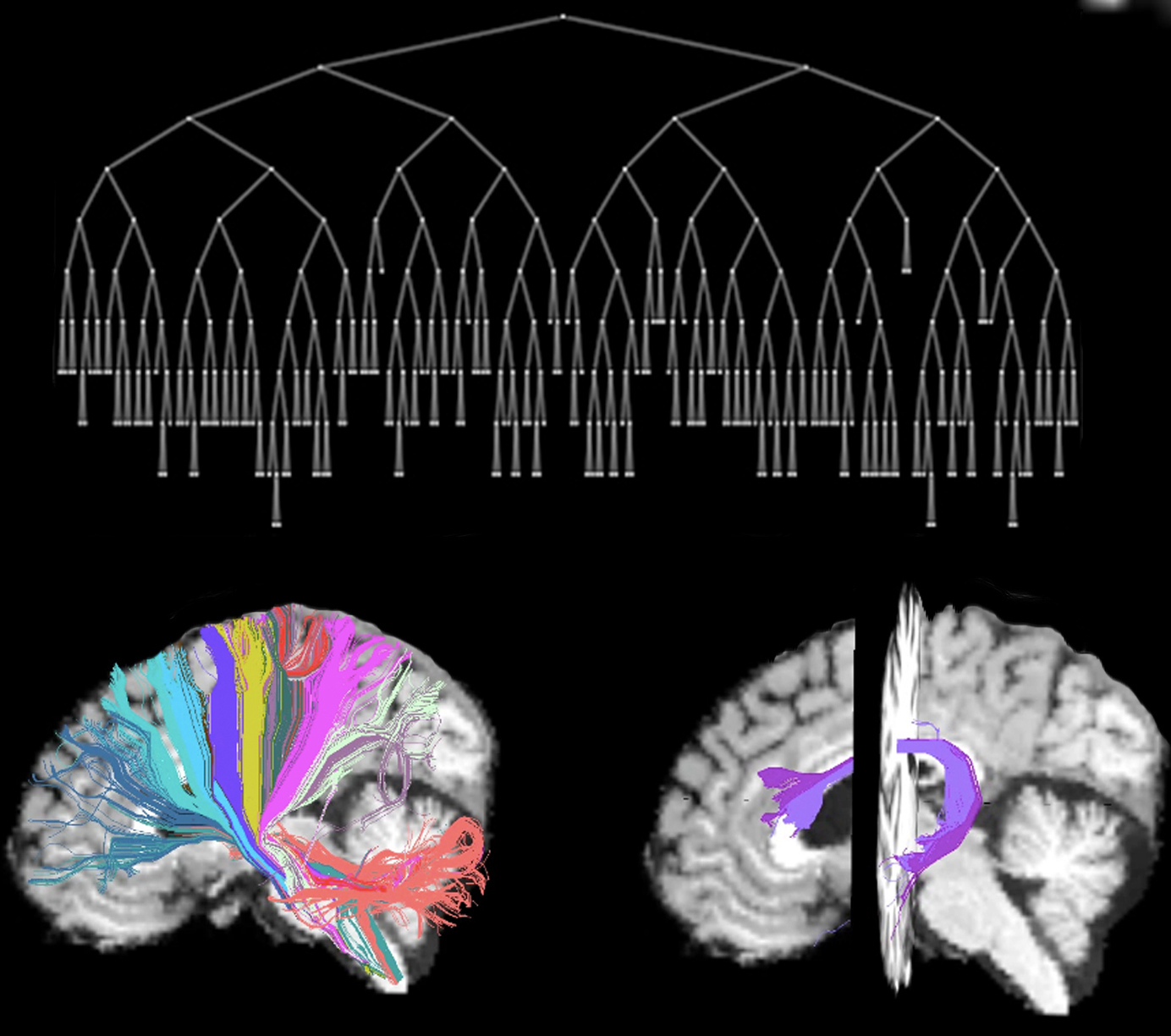
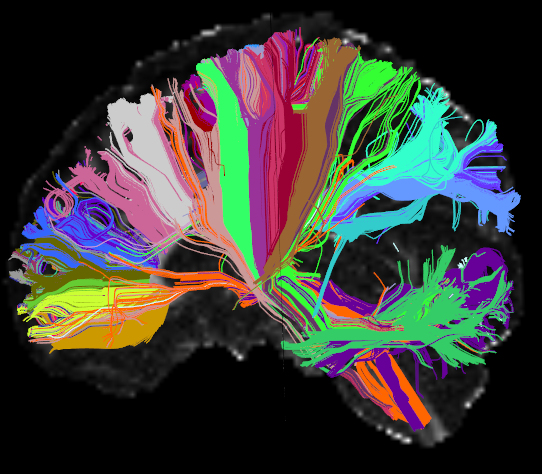
AnatomiCuts is an unsupervised hierarchical clustering that uses an anatomical similarity metric.
Contents
Inputs
- streamlines file (*.trk)
- a segmentation image, preferable including cortical and subcortical parcellation with white matter segmentation based on neighboring regions (wmparc.mgz or wm2009parc.mgz).
It is important to ensure that the streamlines and the segmentation are in the same space. To validate this it is recommended to visualize the streamline file and the segmentation file using both Freeview and Trackvis, since Freeview does not take into account RAS orientation, and Trackvis does not take into account the image offset.
Filtering streamlines
Sometimes deterministic tractography can generate spurious streamlines outside the brain. To exclude them we can use streamlineFilter. It is recommended to remove the short streamlines that can prematurely end during tractography. It is also possible to remove the ushape streamlines.
Running AnatomiCuts
AnatomiCuts -s segmentation.nii.gz -f streamlines.vtk -labels -o outputFolder <options>
Where
-s segmentation file such as wm2009parc.nii.gz
-f streamline file in trk format.
-c number of clusters
-s segmentation file such as wm2009parc.nii.gz
-f streamline file in trk format.
-c number of clusters. The default is 200.
-n number of points to be used per streamline. The default is 10.
-e number of streamlines to be used for the eigendecomposition of normalized cuts algorithm. The default is 500. Using less number of streamlines will fasten substantially the algorithm at the expense of accuracy. For a quick test 50 could be ok, but in practice, less than 300 is not recommended.
-d directional neighbors to be used, the default is "a" all (26), diagonal "d" is 14 and straight "s" is 6.
-n number of points to be used per streamline. The default is 10.
-e number of streamlines to be used for the eigendecomposition of normalized cuts algorithm. The default is 500. Using less number of streamlines will fasten substantially the algorithm at the expense of accuracy. For a quick test 50 could be ok, but in practice, less than 300 is not recommended.
-d directional neighbors to be used, the default is "a" all (26), diagonal "d" is 14 and straight "s" is 6.
Finding corresponding clusters across subjects
You can find one-to-one correspondences using AnatomiCuts_correspondences. .
References
AnatomiCuts: Hierarchical clustering of tractography streamlines based on anatomical similarity. Siless V., Chang K., Fischl B., Yendiki A.. NeuroImage 2018.