|
Size: 2749
Comment:
|
Size: 3157
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 3: | Line 3: |
| The 'Cortical Hubs' (CHubs) are regions-of-interest defined by cortical thickness differences between older controls and individuals classified as having AD, created using the [[OASIS]] data set. These ROIS were used in the study [[https://surfer.nmr.mgh.harvard.edu/pub/articles/Sabuncu2011_Archives_InPress.pdf|The Dynamics of Cortical and Hippocampal Atrophy in Alzheimer Disease, Sabuncu et al.]]. See also: [[https://surfer.nmr.mgh.harvard.edu/pub/articles/Sabuncu_ArchivesOfNeurology_2011_Supplemental.pdf|Supplemental]] and the snapshot of a figure from the paper below. | The 'Cortical Hubs' (CHubs) are regions-of-interest defined by cortical thickness differences between older controls and individuals classified as having AD, created using the [[JenniNotes/Oasis|OASIS]] data set. These ROIs were used in the study [[https://surfer.nmr.mgh.harvard.edu/pub/articles/Sabuncu2011_Archives_InPress.pdf|The Dynamics of Cortical and Hippocampal Atrophy in Alzheimer Disease, Sabuncu et al.]]. See also: [[https://surfer.nmr.mgh.harvard.edu/pub/articles/Sabuncu_ArchivesOfNeurology_2011_Supplemental.pdf|Supplemental]], and below is a snapshot of a figure from the paper. |
| Line 24: | Line 24: |
| To map these labels to your subject set (defined in SUBJECTS): | To map these labels to your subject set (which you would define in a SUBJECTS csh shell var), first you will need to symlink the fsaverage subject into your own subjects dir: {{{ cd $SUBJECTS_DIR ln -s $FREESURFER_HOME/subjects/fsaverage }}} Confirm that you see 16 .label files and two .annot files: {{{ ls $SUBJECTS_DIR/fsaverage/label/*chubs* }}} Now you can do the mapping: |
| Line 50: | Line 61: |
| Snaphot from the Sabuncu paper: | |
| Line 53: | Line 65: |
| * While the paper says these are from the [[OASIS]] data set, they are actually from an earlier data set created at Washington University which later was included in the OASIS data set. The data set used to create these CHubs is found in the NMR Center here: | * While the paper says these are from the [[JenniNotes/Oasis|OASIS]] data set, they are actually from an earlier data set created at Washington University whose subjects were later included in the OASIS data set. The data set used to create these CHubs is found in the NMR Center here: |
| Line 65: | Line 77: |
| These are a bunch of scripts (in the /scripts dir) which performed that mappings... see *chubs* files. | These are a bunch of scripts (in the /scripts dir) which performed those mappings... see *chubs* files. |
Cortical Hubs
The 'Cortical Hubs' (CHubs) are regions-of-interest defined by cortical thickness differences between older controls and individuals classified as having AD, created using the OASIS data set. These ROIs were used in the study The Dynamics of Cortical and Hippocampal Atrophy in Alzheimer Disease, Sabuncu et al.. See also: Supplemental, and below is a snapshot of a figure from the paper.
Freesurfer v5.2 (and v5.1 local) contains these ROIs as labels, found in the directory $FREESURFER_HOME/subjects/fsaverage/label. There are eight:
FS label name |
name used in paper |
?h.oasis.chubs.ifc |
inferior frontal cortex |
?h.oasis.chubs.ipc |
inferior parietal cortex |
?h.oasis.chubs.ips |
inferior parietal sulcus |
?h.oasis.chubs.lateraltemporal |
lateral temporal cortex |
?h.oasis.chubs.medialpfc |
not used in paper |
?h.oasis.chubs.mtl |
medial temporal lobe (eg. entorhinal cortex) |
?h.oasis.chubs.retrosplenial |
posterior cingulate |
?h.oasis.chubs.tp |
temporalpolar cortex |
These labels are also combined into .annot files: ?h.oasis.chubs.annot
To view these on the fsaverage rh surface:
tksurfer fsaverage rh inflated -annotation oasis.chubs
To map these labels to your subject set (which you would define in a SUBJECTS csh shell var), first you will need to symlink the fsaverage subject into your own subjects dir:
cd $SUBJECTS_DIR ln -s $FREESURFER_HOME/subjects/fsaverage
Confirm that you see 16 .label files and two .annot files:
ls $SUBJECTS_DIR/fsaverage/label/*chubs*
Now you can do the mapping:
foreach s ($SUBJECTS)
echo $s
foreach h (lh rh)
foreach l (oasis.chubs.ifc \
.oasis.chubs.ipc \
.oasis.chubs.ips \
.oasis.chubs.lateraltemporal \
.oasis.chubs.mtl \
.oasis.chubs.retrosplenial \
.oasis.chubs.tp)
set cmd=(mri_label2label \
--srclabel fsaverage/label/${h}${l} \
--srcsubject fsaverage \
--trgsubject $s \
--trglabel $s/label/${h}${l} \
--regmethod surface \
--hemi $h)
echo $cmd
$cmd
end
end
endSnaphot from the Sabuncu paper: 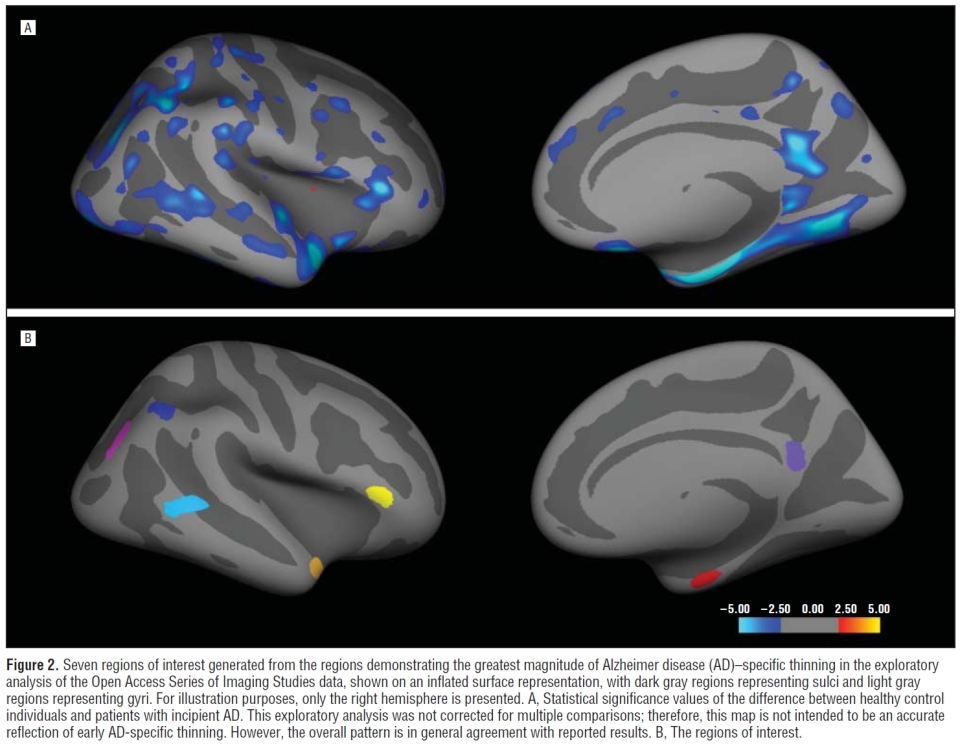
Notes:
While the paper says these are from the OASIS data set, they are actually from an earlier data set created at Washington University whose subjects were later included in the OASIS data set. The data set used to create these CHubs is found in the NMR Center here:
/autofs/cluster/con_008/users/washu
- The CHubs ROIs were mapped to the ADNI data set. The ADNI data set is found here:
/autofs/cluster/con_009/users/ADNI
These are a bunch of scripts (in the /scripts dir) which performed those mappings... see *chubs* files.
