Editing a Recon
Below, we ask you to look at several subjects with various errors that require manual intervention. Open each subject and try to identify the problem on your own. For each subject, there is a link to instructions on how to fix the problem. While you are trying to identify the problems with the subjects listed below you may find it helpful to refer back to the previous exercise, or open your own instance of the subject good_output to compare these subjects to a good example.
Edits to the wm volume
Now, take a look at the next subject, wm1_edits_before.
freeview -v wm1_edits_before/mri/wm.mgz:colormap=heat:opacity=0.4 wm1_edits_before/mri/brainmask.mgz -f wm1_edits_before/surf/lh.white:edgecolor=blue wm1_edits_before/surf/lh.pial:edgecolor=red wm1_edits_before/surf/rh.white:edgecolor=blue wm1_edits_before/surf/rh.pial:edgecolor=red
Again, this will bring up the brainmask.mgz volume, the wm.mgz volume, and the surfaces for both hemispheres.
Click in the main viewing area and use Alt-C to flip between the wm.mgz and brainmask.mgz on top of each other.
with the wm.mgz loaded on top of the brainmask.mgz volume, it should look like this: 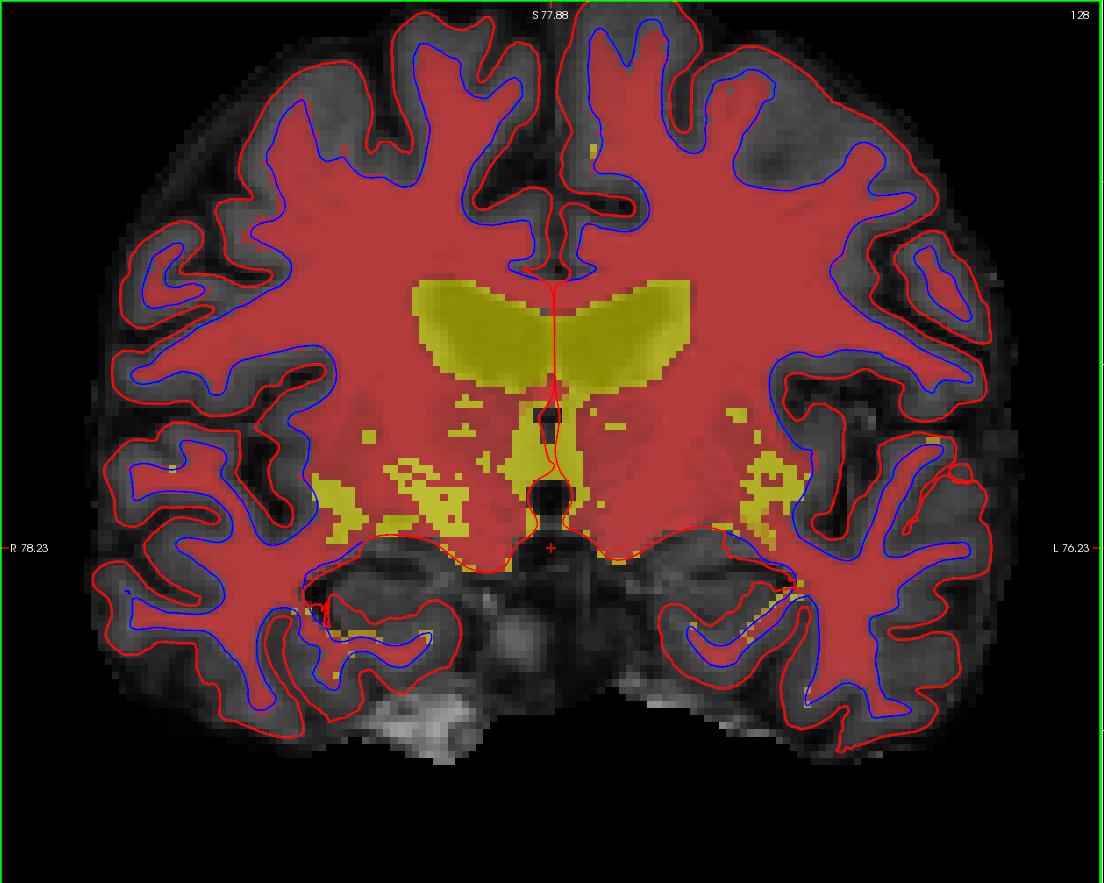
In your second and third terminal window, if not already open, open the surfaces in tksurfer:
tksurfer wm1_edits_before lh inflated
tksurfer wm1_edits_before rh inflated
This will open the inflated surfaces for both hemispheres in tksurfer.
The trouble with this subject has occurred during the white matter segmentation step. Check the surfaces in the T1 volume to find the spot that does not match the actual gray/white boundary. If you load the wm.mgz volume as the aux volume you will see a hole in this area. If you are looking in tksurfer you will see a hole or dimple in the inflated surface. You can use the "save point" "goto point" commands to find this spot in the volume.
Using the Page Up and Page Down keys, you can scroll towards the anterior portion of the brain until you find an area in the right hemisphere where the white surface (blue line) does not follow the surface of the brain, but in fact cuts into it. This geometric innacuracy is cause by a lesion where the white matter has been marked as non-white matter. 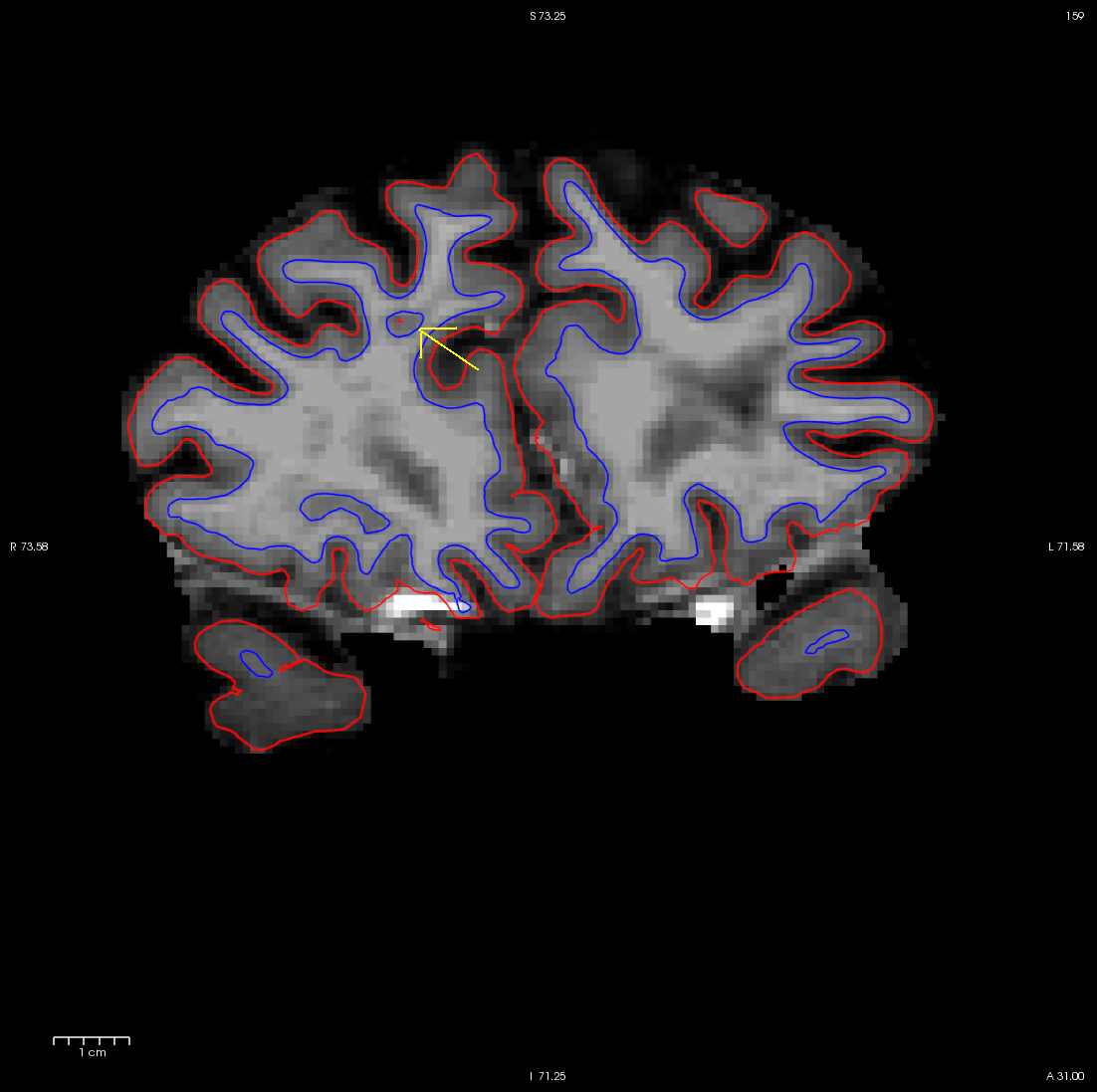
use Alt-C to have the wm.mgz volume loaded on top and you can see that this area has been left out of the wm volume completely.
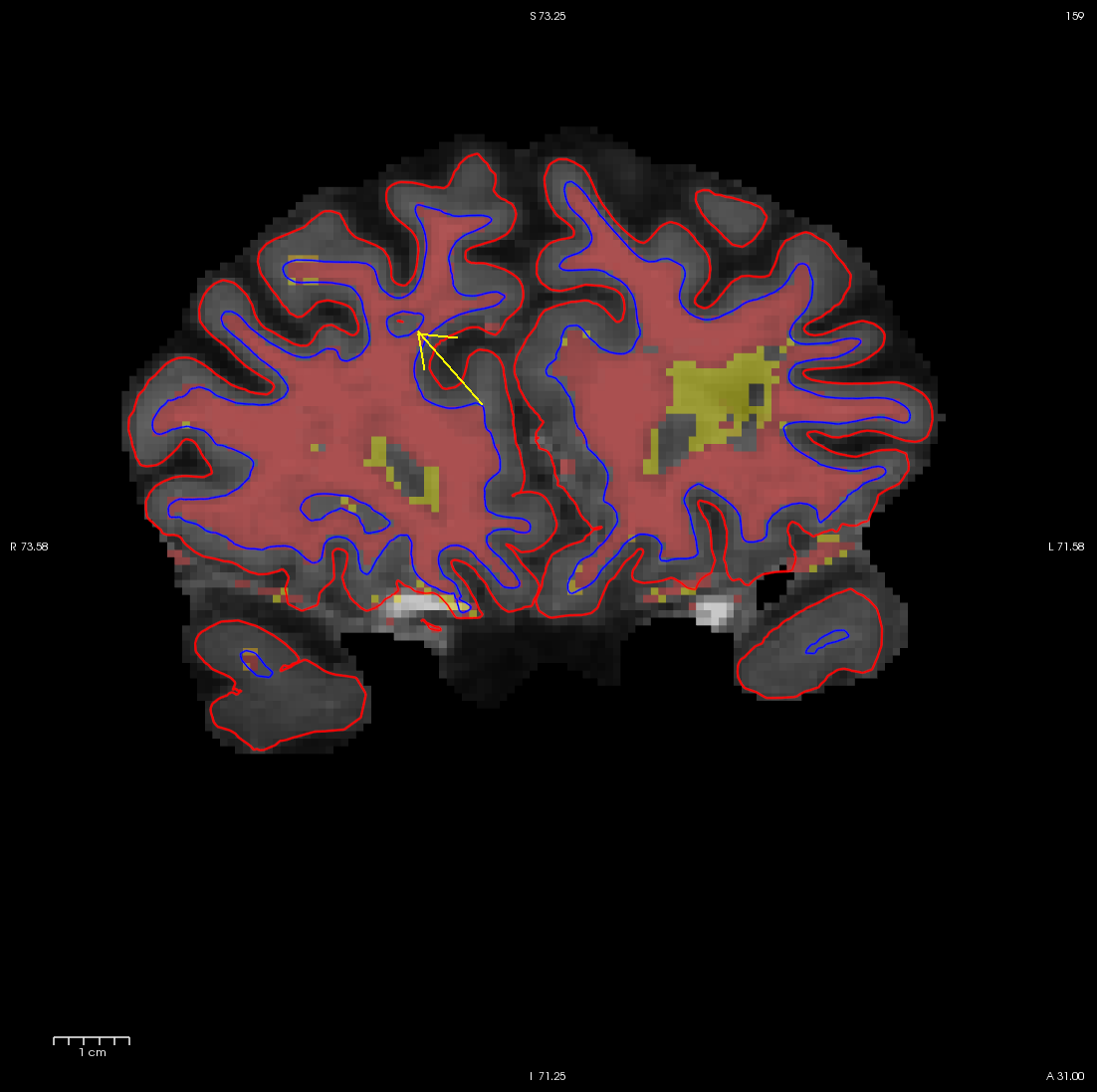
You can see this as a dimple or hole on the inflated surface in tksurfer too (see next image). If not already open, in a second terminal window, open the rh surface in tksurfer and you can see the dimple or hole on the medial side:
tksurfer wm1_edits_before rh inflated
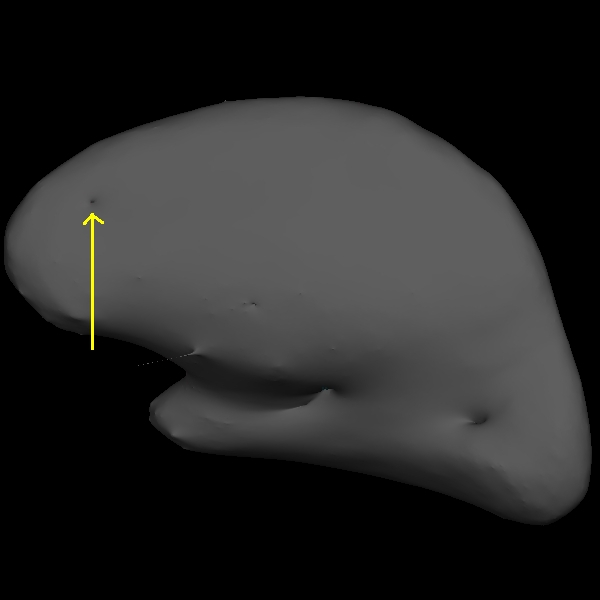
To fix this problem you will need to fill in the missing voxels in the wm.mgz volume. First, it's a good idea to use the page up and down keys to scroll through the individual slices in the volume until you get a good idea of where the problem starts and ends. You will want to start filling in voxels when the inaccuracy appears, and keep filling them in slice by slice until the problem is no longer visible. The coronal view and a brush radius of one or two are good settings for painting in voxels. Zoom In and Out either by using the scroll button on your mouse or holding down shift and the up or down keys.
To begin editing voxels, make sure the wm.mgz is highlighted in the volume menu, and on top on the brainmask.mgz. click on the edit voxels option (  ) and enter 255 into the brush value box. your menu should look like this:
) and enter 255 into the brush value box. your menu should look like this:
in the Voxel edit menu that pops up, click on the first icon "freehand" and select a Brush size of 1 or 2. Make sure the Reference is the wm volume. This menu should look like this:
![]() You can also compare the original data to a corrected and rerun version, wm1_edits_after.
You can also compare the original data to a corrected and rerun version, wm1_edits_after.
To view this corrected subject (which was corrected by following the detailed instructions provided to you):
tkmedit wm1_edits_after brainmask.mgz -aux wm.mgz -surfs
