About
This page describes how to perform seed-based functional connectivity (FC) analysis in FSFAST. This is an extension of the task-based analysis for which there is much more documentation. It may be worth your time to study some of the preprocessing and task-based analysis as found in FS-FAST powerpoint and the FS-FAST tutorial.
*STEP 1: Unpack Data into the FSFAST Hierarchy using dcmunpack (run with -help for more documentation):
Sample cmd:
dcmunpack -src dicomdir -targ sessionid -fsfast -run 3 bold nii.gz f.nii.gz -run 4 bold nii.gz f.nii.gz
In this sample command...
- Have all fMRI dicoms for this subject in the dicomdir folder or subfolders under this folder
- Arguement for "-targ" specifies output directory here called "sessionid". This should be unique to the subject (and visit if longitudinal). This is called the session folder.
- -run 3 bold nii.gz f.nii.gz will unpack run 3 fmri to sessionid/bold/003/f.nii.gz
- To get a list of runs, run dcmunpack -src dicomdir/subject/ALLDICOMS
- Use "-fsfast" to generate fsfast hierarchy shown in the image below
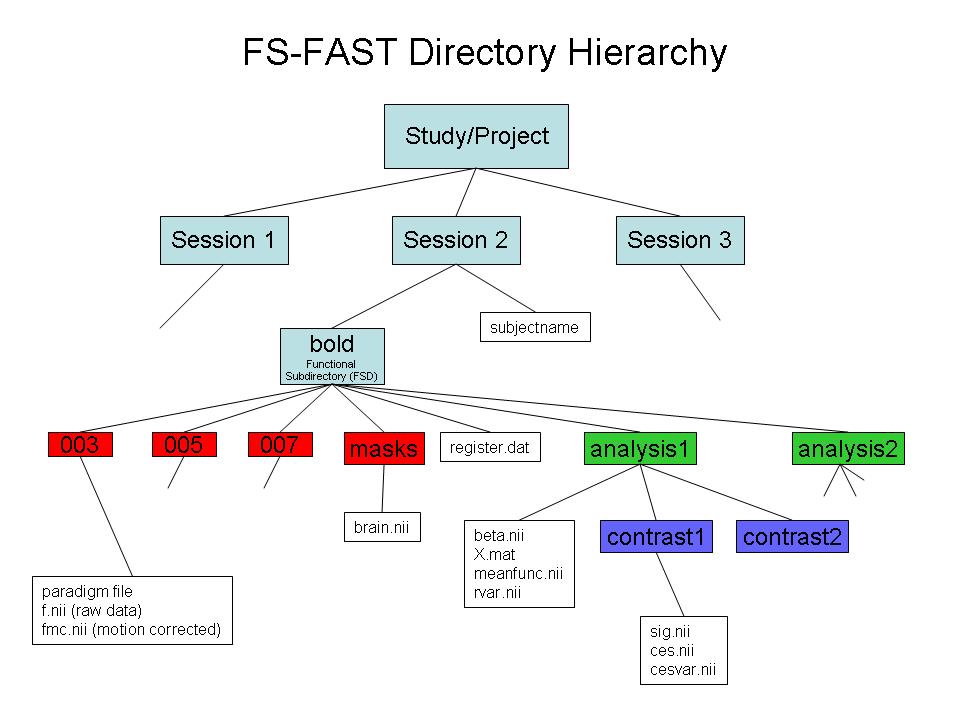
*STEP 2: Link to FreeSurfer anatomical analysis. This is done by creating a text file called sessionid/subjectname with the name of the FreeSurfer anatomical folder as created with recon-all and found in $SUBJECTS_DIR.
*STEP 3: Pre-process your bold data using preproc-sess preproc-sess
Sample cmd:
preproc-sess -s sessionid -fwhm 10 -surface fsaverage lhrh
By default this will do motion correction, masking, registration to the anatomical, sampling to the surface, and surface smoothing. The sampling is done onto the surface of the lh and rh hemispheres of fsaverage. Note that eventhough the time series data are sampled onto fsaverage, the FC seeds are derived from the indvidual anatomy as shown below.
*STEP 4:
There are two methods of deriving a seed region:
1) To use a full-size Freesurfer parcellation from aparc+aseg.mgz, continue with STEP 5 on this page.
2) To split the full Freesurfer parcellation into multiple seeds ("split parcellation"), follow the additional steps here - and resume with step 5 on this page...
*STEP 5: Use fcseed-config to configure the parameters you wish to pass to your connectivity analysis.
Sample command: fcseed-config -segid 1010 -fcname mean.L_Posteriorcingulate.dat -fsd bold -mean -cfg mean.L_Posteriorcingulate.config
This example will use the FreeSurfer cortical segmentation for the left posterior cingulate (segID: 1010) as defined for this individual. For seed regions, we recommend generating the mean signal timecourse by using "-mean". Note that this does not perform any analysis, it just creates a text file with the configuration.
*STEP 5: Pass the config text file to fcseed-sess to generate time-course information for your chosen seed region (or for nuisance variable signal).
fcseed-sess -s sessionid -cfg mean.L_Posteriorcingulate.config
Sample cmd (mean waveforms for nuisance regressors, but PCA also possible with -pca):
for white matter:
- fcseed-config -wm -fcname wm.dat -fsd bold -cfg wm.config
- fcseed-sess -s sessionid -cfg wm.config
for ventricles + CSF:
- fcseed-config -vcsf -fcname vcsf.dat -fsd bold -mean -cfg vcsf.config
fcseed-sess -s <session> -cfg vcsf.config
- NOTE: Once a config file is created it may be used for multiple sessions
*STEP 5: Use mkanalysis-sess to setup an analysis for your FC data
Sample cmd:
mkanalysis-sess -a <analysisname>
-surface fsaverage <hemi> -notask -taskreg mean.L_Posteriorcingulate.dat 1 -nuisreg vcsfreg.dat 1 -nuisreg wmreg.dat 1 -nuisreg global.waveform.dat 1 -fwhm 5 -fsd bold -TR <TR> -mcextreg -polyfit 5 -nskip 4
Note: if you do not want to regress out the global signal, then do not include it in the mkanalysis-sess command.
*STEP 6: Use selxavg3-sess to run the subject-level analysis outlined by the above mkanalysis-sess cmd.
selxavg3-sess -s <session> -a <analysisname>
*STEP 7: Choose the contrast file (generated in each session's contrast directory) that you wish to analyze on a group level:
- # ces.mgz - contrast effect size (contrast matrix * regression coef) # cesvar.mgz - variance of contrast effect size # sig.mgz - significance map (-log10(p)) # pcc.mgz - partial correlation coefficient map
*STEP 8: To continue with a group-level analysis, try one of the methods below:
- Method 1:
- create fsgd file containing all sessions of interest
Concatenate contrast files using mri_concat
Run group analysis using mri_glmfit
Concatenate with isxconcat-sess
Run group analysis using mri_glmfit
