| Slideshow ^ |< << Slide 3 of 14 >> >| |
You will notice several subdirectories and a text file in Bert's directory, and that they are already in a structure called the Sessions Format. If you unpack subject data directly from your facility's scanner, the automatically created subdirectories and files might not conform to the format required by FsFast, i.e. the Sessions Format. You may need to manually arrange and/or rename them, to ensure that they conform to the Sessions Format. Further details are provided in the next step. For now, it is a good idea to familiarize yourself with the FsFast's Sessions Format.
The Sessions Format
The Sessions Format is a structure that demands a certain file and directory naming convention for the scanning acquisitions (sessions). There is some flexibility in how you name the individual files and directories in this Format, but the naming of the sessions must conform exactly to the convention outlined here in order for FsFast to work. New files and directories will be automatically added to this structure during each stage of FsFast processing. Here is a look at Bert's Sessions Format:
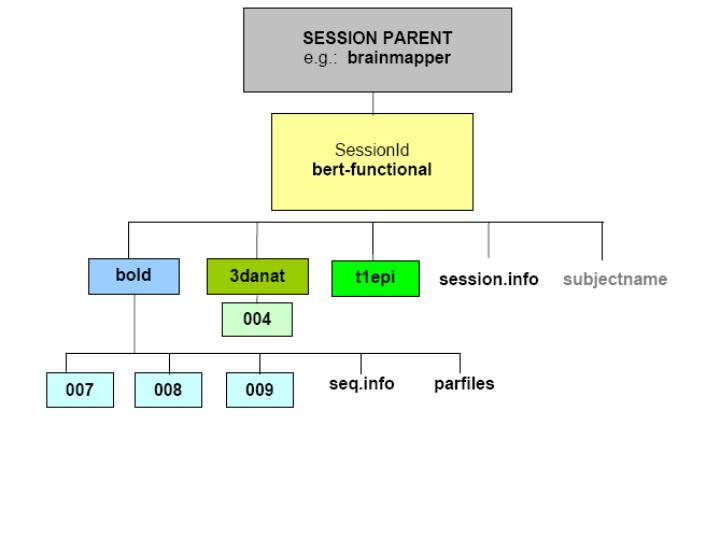
SESSION PARENT: This is the directory to where you unpack subjects' individual session directories, e.g., 'bert-functional.' You can call it whatever you like, e.g., 'brainmapper.'
SessionId: This directory name identifies the session for a particular subject. It contains the MRI or fMRI scan, or series of scans, carried out on that particular subject on a particular occasion. The SessionID is also known simply as the subject's 'session directory.' For this tutorial you will have only one SessionID (Bert's) in your Session Parent.
bold: This is the functional subdirectory. It contains the individual run(s) of functional MRI data; each of which must be named by three-digit, zero-padded numbers (e.g., 007, 008, 009) in order for FsFast to recognize them as functional data directories. (Although not applicable in this case, if this directory were named something other than 'bold,' FsFast would require modifications to further steps in the processing stream. These variations are described in the FsFastGuide.)
007/008/009: Under the bold directory, these are the subdirectories where each functional volume (or 'run' ) is stored, in bfile format (i.e., bshorts) with an 'f' stem.
3danat: This directory contains any raw structural MR data collected during the session, and stores it in COR format. Each volume/structural run should be in an individual subdirectory.
004: Under 3danat, this subdirectory contains only anatomical data from the fast structural scan (i.e., 004), acquired on the same date as the functional session. The anatomical data from this fast scan will later be used for registration of the functional data with Bert's reconstructed anatomical data. Bert's anatomical data (acquired with a different scan protocol) that were used for the anatomical reconstruction would initially appear here also, but now have a distinctive directory structure of their own, one that is automatically created in the FreeSurfer reconstruction.
t1epi: This directory contains the t1-weighted EPI image data collected during the session, whose slice prescription must be identical to that of the functional data. Bert's actual t1epi volume is stored in a subdirectory called 004 in bshort format, with a stem called 'f.'
session.info: This text file contains general information about the subject (e.g., DOB, gender, console name, population, study date, etc.).
subjectname: This text file contains the subject identifier string (i.e., the name of the directory containing the subject's anatomical directory, e.g., 'bert-anatomical'), and nothing else. You will create this text file in a subsequent step.
seq.info: This text file contains details such as the scanning protocol used during the scan, date of scan, etc.
parfiles: These paradigm files give information about the stimulus schedule for each run, that is, details about which stimulus was presented at which time during the functional run. You should have one paradigm file for each functional run. The parfiles are created by the FsFast program optseq2.
| Slideshow ^ |< << Slide 3 of 14 >> >| |
