| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| [wiki:Self:FsTutorial top] | [wiki:Self:FsTutorial previous] | [wiki:Self:FsTutorial/Visualization next] | [[FsTutorial|top]] | [[FsTutorial|previous]] | [[FsTutorial/Visualization|next]] |
| Line 6: | Line 6: |
| '''Please see the new group analysis tutorial, using Qdec. Click''' [wiki:Self:FsTutorial/QdecGroupAnalysis here] | '''Please see the new group analysis tutorial, using Qdec. Click''' [[FsTutorial/QdecGroupAnalysis|here]] |
| Line 11: | Line 11: |
| In this tutorial, you will learn how to perform statistical analysis of group surface-based data, including:[[BR]] | In this tutorial, you will learn how to perform statistical analysis of group surface-based data, including:<<BR>> |
| Line 13: | Line 13: |
|
*Constructing a FreeSurfer Group Descriptor File (FSGD)[[BR]] *Preprocessing the group data[[BR]] *Constructing the design matrix[[BR]] *Constructing contrast matrices to test hypotheses[[BR]] *Correcting for multiple comparisons[[BR]] |
*Constructing a FreeSurfer Group Descriptor File (FSGD)<<BR>> *Preprocessing the group data<<BR>> *Constructing the design matrix<<BR>> *Constructing contrast matrices to test hypotheses<<BR>> *Correcting for multiple comparisons<<BR>> |
| Line 37: | Line 37: |
| (Note: the data in the group_study directory is not required to complete the tutorial. However, [wiki:Self:FsTutorial/Data it is available for download] if you wish to pursue your own group analysis.) | (Note: the data in the group_study directory is not required to complete the tutorial. However, [[FsTutorial/Data|it is available for download]] if you wish to pursue your own group analysis.) |
| Line 40: | Line 40: |
| Linear modeling describes the observed data as a linear combination of explanatory factors plus noise, and determines how well that description explains the data being analyzed. In order to understand how to perform group analysis in FreeSurfer, you need to understand the general linear model (GLM) and how to construct a GLM in matrix notation. You can click [wiki:Self:FsTutorial/GlmReview here] for a review of this material. The notation we use here is:'''y=X*beta''', where '''y''' is the vector observed data (e.g., thicknesses for each subject at a vertex), '''X''' is the known design matrix (e.g., gender, age), and '''beta''' is the vector of unknown parameter estimates (PEs). The interpretation of the PEs will depend upon how '''X''' is constructed. For example, they could be interpeted as a slope indicating the change of thickness with age. The analysis/estimation is then the process of computing '''beta''' given the data '''y''' and the design matrix '''X'''. A Null Hypothesis (H0) is constructed with a contract matrix '''C'''. Inferences are drawn by testing whether the value '''gamma=Cb''' is zero. | Linear modeling describes the observed data as a linear combination of explanatory factors plus noise, and determines how well that description explains the data being analyzed. In order to understand how to perform group analysis in FreeSurfer, you need to understand the general linear model (GLM) and how to construct a GLM in matrix notation. You can click [[FsTutorial/GlmReview|here]] for a review of this material. The notation we use here is:'''y=X*beta''', where '''y''' is the vector observed data (e.g., thicknesses for each subject at a vertex), '''X''' is the known design matrix (e.g., gender, age), and '''beta''' is the vector of unknown parameter estimates (PEs). The interpretation of the PEs will depend upon how '''X''' is constructed. For example, they could be interpeted as a slope indicating the change of thickness with age. The analysis/estimation is then the process of computing '''beta''' given the data '''y''' and the design matrix '''X'''. A Null Hypothesis (H0) is constructed with a contract matrix '''C'''. Inferences are drawn by testing whether the value '''gamma=Cb''' is zero. |
| Line 50: | Line 50: |
| The following study variables correspond to all the subjects found in {{{$FREESURFER_HOME/subjects/buckner_data/tutorial_subjs/group_analysis_tutorial/}}} and are presented in a spreadsheet. You'll need to compose an FSGD file with the appropriate variables in order to specify a design matrix that can be used to examine the relationship between a subject's age and cortical thickness. You can read more about the rules for creating an FSGD file [wiki:Self:FsTutorial/CreateFsgdFile here]. To get started, open the text editor of your choice and add individual tags by following the general example and rules described [wiki:Self:FsTutorial/CreateFsgdFile here]. In general, it's a good idea to name the file something intuitive, such as my_gender_age_fsgd.txt. To create your FSGD file you first need to change to the directory you'd like to create the file in: | The following study variables correspond to all the subjects found in {{{$FREESURFER_HOME/subjects/buckner_data/tutorial_subjs/group_analysis_tutorial/}}} and are presented in a spreadsheet. You'll need to compose an FSGD file with the appropriate variables in order to specify a design matrix that can be used to examine the relationship between a subject's age and cortical thickness. You can read more about the rules for creating an FSGD file [[FsTutorial/CreateFsgdFile|here]]. To get started, open the text editor of your choice and add individual tags by following the general example and rules described [[FsTutorial/CreateFsgdFile|here]]. In general, it's a good idea to name the file something intuitive, such as my_gender_age_fsgd.txt. To create your FSGD file you first need to change to the directory you'd like to create the file in: |
| Line 56: | Line 56: |
| attachment:demoTable2.jpg | {{attachment:demoTable2.jpg}} |
| Line 64: | Line 64: |
| A correct FSGD file is presented [wiki:Self:FsTutorial/CorrectFsgdFile here] for comparison. It is needed for a later exercise, so create it now if you have not already done so. | A correct FSGD file is presented [[FsTutorial/CorrectFsgdFile|here]] for comparison. It is needed for a later exercise, so create it now if you have not already done so. |
| Line 81: | Line 81: |
| Please click [wiki:Self:FsTutorial/DesignMatrix here] for further explanation of the design matrix and how it is used with the FSGD file. | Please click [[FsTutorial/DesignMatrix|here]] for further explanation of the design matrix and how it is used with the FSGD file. |
| Line 117: | Line 117: |
| mri_glmfit performs the general linear model (GLM) analysis on the volume or the surface. Options include simulation for correction for multiple comparisons, weighted LMS, variance smoothing, PCA/SVD analysis of residuals, per-voxel design matrices, and 'self' regressors. This program performs both the estimation and inference. The framework for testing specific hypotheses is specified in the form of a contrast vector. For each comparison you want to run you will need to design a separate contrast vector. For information on how to set up a contrast vector please click [wiki:Self:FsTutorial/CreateContrastVectors here]. | mri_glmfit performs the general linear model (GLM) analysis on the volume or the surface. Options include simulation for correction for multiple comparisons, weighted LMS, variance smoothing, PCA/SVD analysis of residuals, per-voxel design matrices, and 'self' regressors. This program performs both the estimation and inference. The framework for testing specific hypotheses is specified in the form of a contrast vector. For each comparison you want to run you will need to design a separate contrast vector. For information on how to set up a contrast vector please click [[FsTutorial/CreateContrastVectors|here]]. |
| Line 129: | Line 129: |
| As an additional exercise, try constructing the contrast vector for testing the same data for difference between males and females independent of age. [wiki:Self:FsTutorial/MaleFemaleContrastSolution Click here for the answer.] | As an additional exercise, try constructing the contrast vector for testing the same data for difference between males and females independent of age. [[FsTutorial/MaleFemaleContrastSolution|Click here for the answer.]] |
| Line 152: | Line 152: |
|
ar1.mgh - spatial AR1 coefficients [[BR]] beta.mgh - all regression coefficients[[BR]] eres.mgh - residual error[[BR]] fsgd.X.mat - final design matrix in matlab format [[BR]] mri_glmfit.log - execution parameters[[BR]] rstd.mgh - residual error stddev (just sqrt of rvar)[[BR]] rvar.mgh - residual error variance[[BR]] Xg.dat - design matrix in text format [[BR]] y.fsgd - fsgd file[[BR]] |
ar1.mgh - spatial AR1 coefficients <<BR>> beta.mgh - all regression coefficients<<BR>> eres.mgh - residual error<<BR>> fsgd.X.mat - final design matrix in matlab format <<BR>> mri_glmfit.log - execution parameters<<BR>> rstd.mgh - residual error stddev (just sqrt of rvar)<<BR>> rvar.mgh - residual error variance<<BR>> Xg.dat - design matrix in text format <<BR>> y.fsgd - fsgd file<<BR>> |
| Line 170: | Line 170: |
|
C.dat - copy of contrast matrix[[BR]] F.mgh - F-ratio[[BR]] gamma.mgh - contrast[[BR]] sig.mgh - significance from F-test (actually -log10(p))[[BR]] |
C.dat - copy of contrast matrix<<BR>> F.mgh - F-ratio<<BR>> gamma.mgh - contrast<<BR>> sig.mgh - significance from F-test (actually -log10(p))<<BR>> |
| Line 195: | Line 195: |
| '''4.1 Simulations''' [[BR]] | '''4.1 Simulations''' <<BR>> |
| Line 204: | Line 204: |
|
(1) perm - perumation, randomly permute rows of X (cf FSL randomise) [[BR]] (2) mc-full - full Monte Carlo simulation, replace input with white gaussian noise, smooth, and analyze. [[BR]] (3) mc-z - z Monte Carlo simulation. Does not actually do analysis, just assume the output is z-distributed (cf ANFI AlphaSim)[[BR]] |
(1) perm - perumation, randomly permute rows of X (cf FSL randomise) <<BR>> (2) mc-full - full Monte Carlo simulation, replace input with white gaussian noise, smooth, and analyze. <<BR>> (3) mc-z - z Monte Carlo simulation. Does not actually do analysis, just assume the output is z-distributed (cf ANFI AlphaSim)<<BR>> |
| Line 248: | Line 248: |
| '''4.2 Clustering''' [[BR]] | '''4.2 Clustering''' <<BR>> |
TUTORIAL LOCATION HAS MOVED
Good news for those of you who are confused by FSGD files, and mri_glmfit commands: There is a new group analysis tool in Freesurfer, Qdec. To accompany this tool there is also a new group analysis tutorial.
Please see the new group analysis tutorial, using Qdec. Click here
FreeSurfer Tutorial: Group Analysis
- To follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names.
In this tutorial, you will learn how to perform statistical analysis of group surface-based data, including:
- Making an average subject from your set of subjects
Constructing a FreeSurfer Group Descriptor File (FSGD)
Preprocessing the group data
Constructing the design matrix
Constructing contrast matrices to test hypotheses
Correcting for multiple comparisons
Assuming that all surface reconstruction has been completed for all subjects in the study, FreeSurfer's mri_glmfit command can be used to perform inter-subject/group averaging and inference on the cortical surface. Mri_glmfit models the data as a linear combination of effects related to variables of interest, confounds and errors, and permits statistical inferences to be made about effects of interest in relation to error variance. It also allows for certain permutation testing and other means for correcting for mutliple comparisons. For group analysis, this technique fits a general linear model (GLM) at each surface vertex to explain the data from all subjects in the study. In this section, a brief overview of linear modeling is presented and mri_glmfit is described for estimating a linear model and testing hypotheses. The modeling overview can be skipped if this material is already familiar. Other software packages have similar types of programs (e.g., FSL's GFEAT).
1.0 Preparing for Group Analysis
For group analysis, you can create an average subject from all the participants in the study. This average will be used as the target subject upon which the results of your group analysis can be output and viewed. To create this average, use make_average_subject. One has already been created for the later exercise, so there is no need to execute this sample command:
make_average_subject --subjects <subj1> <subj2> ...
The default behavior of this script is to create a subject in the $SUBJECTS_DIR named 'average' using each subjects talairach.xfm transform. This behavior can be modified on the command line. You can specify --out your_named_average to change the name of the average subject and --xform talairach.lta (or talairach.m3z) to specify the use of one of the other transforms.
The average subject is created using the processed volumes and surfaces from the set of subjects you specify following the --subjects flag. The make_average_subject command executes both the make_average_volume and make_average_surface subscripts for you.
The distributed example of an average subject can be found in $FREESURFER_HOME/subjects/buckner_data/tutorial_subjs/group_analysis_tutorial called fsaverage
This average subject was created using the volumes and surfaces of subjects in:
$FREESURFER_HOME/subjects/buckner_data/group_study
(Note: the data in the group_study directory is not required to complete the tutorial. However, it is available for download if you wish to pursue your own group analysis.)
2.0 Linear Modeling overview
Linear modeling describes the observed data as a linear combination of explanatory factors plus noise, and determines how well that description explains the data being analyzed. In order to understand how to perform group analysis in FreeSurfer, you need to understand the general linear model (GLM) and how to construct a GLM in matrix notation. You can click here for a review of this material. The notation we use here is:y=X*beta, where y is the vector observed data (e.g., thicknesses for each subject at a vertex), X is the known design matrix (e.g., gender, age), and beta is the vector of unknown parameter estimates (PEs). The interpretation of the PEs will depend upon how X is constructed. For example, they could be interpeted as a slope indicating the change of thickness with age. The analysis/estimation is then the process of computing beta given the data y and the design matrix X. A Null Hypothesis (H0) is constructed with a contract matrix C. Inferences are drawn by testing whether the value gamma=Cb is zero.
3.0 Using mri_glmfit for estimating the linear model and hypothesis testing
As stated earlier, mri_glmfit performs inter-subject/group averaging and inference on the surface by fitting a linear model at each vertex. The model consists of subject parameters (e.g., age, gender, etc). The model is the same across all vertices, though the fit will probably be different at each vertex. To specify the model, a design matrix that represents the GLM must be created.
Create an FSGD file
The FreeSurfer Group Descriptor File (FSGDF) provides a way to describe a group of subjects and their accompanying data. This can include class membership and other continuous variables, for example gender or age. When it exists, the FSGDF is used by mri_glmfit, tksurfer and tkmedit. The FSGDF is more than just a way to specify the design matrix. It also keeps track of group membership and covariate definitions. This information is then used to help visualize the results. This is not possible when only a design matrix is used.
The following study variables correspond to all the subjects found in $FREESURFER_HOME/subjects/buckner_data/tutorial_subjs/group_analysis_tutorial/ and are presented in a spreadsheet. You'll need to compose an FSGD file with the appropriate variables in order to specify a design matrix that can be used to examine the relationship between a subject's age and cortical thickness. You can read more about the rules for creating an FSGD file here. To get started, open the text editor of your choice and add individual tags by following the general example and rules described here. In general, it's a good idea to name the file something intuitive, such as my_gender_age_fsgd.txt. To create your FSGD file you first need to change to the directory you'd like to create the file in:
cd $FREESURFER_HOME/subjects/buckner_data/tutorial_subjs/group_analysis_tutorial/stats
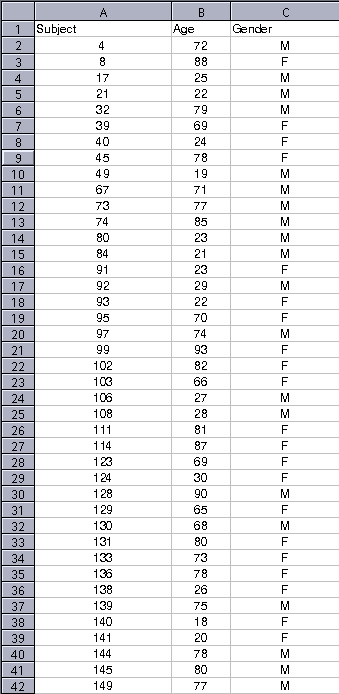
Upon completion, save the FSGD file, which can be viewed in the xterm by typing:
cat my_gender_age_fsgd.txt
in the directory where it has been saved.
A correct FSGD file is presented here for comparison. It is needed for a later exercise, so create it now if you have not already done so.
Creating a Design Matrix
The FSGDF is specified in the command-line for mri_glmfit with the option --fsgd fname <gd2mtx> where gd2mtx is the method by which the group description is converted into a design matrix. Legal values for gd2mtx are:
doss (different offset, same slope): this will create a design matrix in which each class has its own offset but forces all classes to have the same slope.
dods (different offset, different slope): this value models each class with its own offset and slope (default).
none: this value is used if neither of the previous models work for your particular analysis. Using this value requires that you specify the design matrix manually.
If you do not specify one of the above methods, dods will be used by default.
Note: It is not necessary to run mri_glmfit now to create the design matrix, as mri_glmfit will create it for you later in this exercise.
Please click here for further explanation of the design matrix and how it is used with the FSGD file.
Preprocessing steps
These are sample commands that you should run for the group analysis. In interest of time these steps have already been run for you, and the output is found in $FREESURFER_HOME/subjects/buckner_data/tutorial_subjs/group_analysis_tutorial/stats
Once an FSGD file is set up and the average subject is made the preprocessing steps can be followed. The first step will use mris_preproc to assemble your data into a single file in the common surface space, average for this example (which is the average that has been made for this particular study). In this step you will have to specify your FSGD file, gender_age.txt here, your target subject, average here, the hemisphere and measure you are using. You will also name the output file - it's a good idea to use a naming convention that will make it obvious what comparison you are working with. Before running the command you want to be sure your SUBJECTS_DIR is set appropriately and you are in the glm directory you wish to run the analyses in:
setenv SUBJECTS_DIR $FREESURFER_HOME/subjects/buckner_data/tutorial_subjs/group_analysis_tutorial cd $FREESURFER_HOME/subjects/buckner_data/tutorial_subjs/group_analysis_tutorial/stats
now you can run the mris_preproc command:
mris_preproc --fsgd gender_age.txt \ --target fsaverage \ --hemi lh \ --meas thickness \ --out lh.gender_age.thickness.mgh
The next step is to do surface smoothing. Smooth input with a Gaussian kernel with the given full-width/half-maximum (fwhm) specified in mm. For all examples to follow we will use a fwhm = 10mm. To do this mri_surf2surf will be used along with the output from mris_preproc and your average subject.
mri_surf2surf --hemi lh \ --s fsaverage \ --sval lh.gender_age.thickness.mgh \ --fwhm 10 \ --tval lh.gender_age.thickness.10.mgh
You can do the surface smoothing as part of the first step with mris_preproc, but if you do it afterwards as a separate step you can smooth to many different levels without having to rebuild the data each time.
mri_glmfit
As stated earlier, these are sample commands that you should run for the group analysis. In interest of time these steps have already been run for you, and the output is found in $FREESURFER_HOME/subjects/buckner_data/tutorial_subjs/group_analysis_tutorial/stats
mri_glmfit performs the general linear model (GLM) analysis on the volume or the surface. Options include simulation for correction for multiple comparisons, weighted LMS, variance smoothing, PCA/SVD analysis of residuals, per-voxel design matrices, and 'self' regressors. This program performs both the estimation and inference. The framework for testing specific hypotheses is specified in the form of a contrast vector. For each comparison you want to run you will need to design a separate contrast vector. For information on how to set up a contrast vector please click here.
To use this in your mri_glmfit command create a file that contains just the one line of your contrast vector using the text editor of your choice. Again, using an appropriate naming convention is a good idea. Make sure this contrast vector has been created for you and is called age.mat:
cd $FREESURFER_HOME/subjects/buckner_data/tutorial_subjs/group_analysis_tutorial/glm cat age.mat
Create age.mat if it does not exist. It should contain this line:
0 0 1
As an additional exercise, try constructing the contrast vector for testing the same data for difference between males and females independent of age. Click here for the answer.
mri_glmfit will take the output from your smoothing step above, your fsgd file, your average subject and the contrast vector as inputs. You will also have to specify a glm directory name, and in this directory all of the outputs will be saved. It is a good idea to use a descriptive name, so you can easily recognize which outputs are in which directory.
mri_glmfit --y lh.gender_age.thickness.10.mgh \ --fsgd gender_age.txt doss\ --glmdir lh.gender_age.glmdir \ --surf average lh \ --C age.mat
The flag --surf is used to specify that the input has a surface geometry from the hemisphere of the given FreeSurfer subject. If --surf is not specified, then mri_glmfit will assume that the data are volume-based and use the geometry as specified in the header to make spatial calculations.
When this command is finished you will have an lh.gender_age.glmdir. There will be a number of output files in this directory, as well as two other directories. If you did an ls in your glmdir there will be:
ar1.mgh eres.mgh mri_glmfit.log rstd.mgh age/ y.fsgd beta.mgh fsgd.X.mat rvar.mgh Xg.dat
the outputs are as follows:
ar1.mgh - spatial AR1 coefficients
beta.mgh - all regression coefficients
eres.mgh - residual error
fsgd.X.mat - final design matrix in matlab format
mri_glmfit.log - execution parameters
rstd.mgh - residual error stddev (just sqrt of rvar)
rvar.mgh - residual error variance
Xg.dat - design matrix in text format
y.fsgd - fsgd file
There will be a subdirectory for each contrast that you specify. The name of the directory will be that of the contrast matrix file (without the .mat extension). The age directory will have the following files:
C.dat F.mgh gamma.mgh sig.mgh
The outputs are as follows:
C.dat - copy of contrast matrix
F.mgh - F-ratio
gamma.mgh - contrast
sig.mgh - significance from F-test (actually -log10(p))
4.0 Using mri_glmfit to correct for multiple comparisons
One method for correcting for multiple comparisons is to perform simulations under the null hypothesis and see how often the value of a statistic from the 'true' analysis is exceeded. This frequency is then interpreted as a p-value which has been corrected for multiple comparisons. This is especially useful with surface-based data as traditional random field theory is harder to implement. This simulator is roughly based on FSLs permuation simulator (randomise) and AFNIs null-z simulator (AlphaSim). Note that FreeSurfer also offers False Discovery Rate (FDR) correction in tkmedit and tksurfer.
The estimation, simulation, and correction are done in three distinct phases:
- Estimation: run the analysis on your data without simulation. Note: at this point you can view your results with FDR thresholding in tksurfer. FDR is often conservative relative to cluster-based thresholding.
- Simulation: run the simulator with the same parameters as the estimation to get the Cluster Simulation Data (CSD).
- Clustering: run mri_surfcluster, passing it the CSD from the simulator and the output of the estimation. These programs will print out clusters along with their p-values.
The estimation step has been described above, using mri_glmfit with no simulation. The simulation step is run using mri_glmfit again, adding in a simulation flag and parameters. If a design is non-orthogonal the permutation simulation can not be run, instead a simple monte carlo simulation can be run. The clustering step is run with mri_surfcluster.
4.1 Simulations
The simulator synthesizes data under the null hypothesis, analyzes, thresholds, clusters, and then keeps track of the number of times clusters of a given size were encounted. The simulation is invoked by calling mri_glmfit and specifying a simulation type and it's associated parameters, with the flag --sim which is to be followed by 4 parameters:
--sim nulltype nsim thresh csdbasename
The first parameter the nulltype, which is the method of generating the null data to be tested. Valid options are:
(1) perm - perumation, randomly permute rows of X (cf FSL randomise)
(2) mc-full - full Monte Carlo simulation, replace input with white gaussian noise, smooth, and analyze.
(3) mc-z - z Monte Carlo simulation. Does not actually do analysis, just assume the output is z-distributed (cf ANFI AlphaSim)
Which one should you use? Permutation makes the fewest assumptions and is fastest, but it requires that the design matrix be orthogonal (e.g., you cannot have continuous variables such as age or IQ). If you cannot use permutation, then a mc-z will be fastest, but the z assumption requires that you have many subjects (on the order of 80). Otherwise, you must run the full MC simulation, which can take quite a while. For both MC simulations, you must supply the smoothness of your data as Full-Width/Half-Max (FWHM) of the residual. This can be obtained from the ResidualFWHM value in y.fsgd in the glm output directory.
The next parameter is nsim which corresponds to the number of simulations to run. If you want to make inferences at the .01 level then you'll need about 10000 iterations. You can run multiple simulations in parallel, if you have multiple processors, to cut down on processing time.
The next parameter is thresh which corresponds to your threshold and is specified as a -log10(pvalue). Eg, for a p-value threshold of .01, use thresh=2.
The last parameter is csdbasename which corresponds to the base name of the file which will store the Cluster Simulation Data (CSD). Each contrast will get it's own file. When running multiple simulations in parallel be sure to use a unique csdbasename for each run.
Here's a sample command to run the mc-full simulation with 10000 iterations and a p-value threshold of .01:
mri_glmfit --y lh.gender_age.thickness.10.mgh \ --glmdir lh.gender_age.glmdir \ --fsgd gender_age.txt doss \ --surf fsaverage lh \ --fwhm 14.517 --C age.mat \ --sim mc-full 10000 2 lh.gender_age.glmdir/csd1
this will create lh.gender_age.glmdir/csd1-age.csd
If you want to split this into multiple runs you could use the following two commands:
mri_glmfit --y lh.gender_age.thickness.10.mgh \ --fsgd gender_age.txt doss\ --fwhm 14.517 \ --glmdir lh.gender_age.glmdir \ --surf fsaverage lh \ --C age.mat \ --sim mc-full 5000 2 lh.gender_age.glmdir/csd1 mri_glmfit --y lh.gender_age.thickness.10.mgh \ --fsgd gender_age.txt doss\ --fwhm 14.517 \ --glmdir lh.gender_age.glmdir \ --surf average lh \ --C age.mat \ --sim mc-full 5000 2 lh.gender_age.glmdir/csd2
which will generate csd1-age.csd and csd2-age.csd
4.2 Clustering
Using the outputs from the estimation step and the simulations, mri_surfcluster (or mri_volcluster) will create two outputs: the summary file with a table of the clusters it found, and an output surface map of the clusters wth the cluster-wise p-value. The sample mri_surfcluster command is:
mri_surfcluster --in lh.gender_age.glmdir/age/sig.mgh \ --csd lh.gender_age.glmdir/csd1-age.csd \ --csd lh.gender_age.glmdir/csd2-age.csd \ --sum lh.gender_age.glmdir/age/sig.cluster.sum \ --cwsig lh.gender_age.glmdir/age/sig.cluster.mgh
you can pass all the CSD files that were created through this command by adding as many --csd as you need.
The surfcluster summary file, sig.cluster.sum, will look like this:
# ClusterNo Max VtxMax Size(mm^2) TalX TalY TalZ CWP CWPLow CWPHi NVtxs 1 1.850 134208 391.63 -28.8 34.0 -8.0 0.00100 0.00060 0.00140 542 2 1.439 18669 0.56 -10.9 9.1 -13.2 0.99960 0.99930 0.99980 1 3 1.366 148132 13.84 -7.4 11.8 -14.1 0.97440 0.97240 0.97640 25
CWP stands for cluster-wise probability which is the probability after correction for multiple comparisons. The CWP column is the nomial p-value. CWPLow and CWPHi are the 90% confidence intervals on the p-value. Each cluster gets its own p-value, which depends upon its size. NVtxs is the number of vertexes in the cluster. The output surface map, sig.cluster.mgh, will be a map of these clusters with their CWP. This can be viewed with:
tksurfer fsaverage lh inflated \ -overlay lh.gender_age.glmdir/age/sig.cluster.mgh
