Making FinalSurf edits
To follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names.
Below is a picture of how the longitudinal results for MR2 looked originally. Create a copy of 'brain.finalsurfs.mgz' named 'brain.finalsurfs.manedit.mgz' with the following command:
cp OAS2_0002_MR2.long.OAS2_0002/mri/brain.finalsurfs.mgz OAS2_0002_MR2.long.OAS2_0002/mri/brain.finalsurfs.manedit.mgz
Open the volume with the following:
freeview -v OAS2_0002_MR2.long.OAS2_0002/mri/brain.finalsurfs.manedit.mgz \
OAS2_0002_MR2.long.OAS2_0002/mri/brainmask.mgz \
OAS2_0002_MR2.long.OAS2_0002/mri/aseg.mgz:colormap=lut:opacity=0.25 \
-f OAS2_0002_MR2.long.OAS2_0002/surf/lh.pial:edgecolor=red \
OAS2_0002_MR2.long.OAS2_0002/surf/rh.pial:edgecolor=red \
OAS2_0002_MR2.long.OAS2_0002/surf/lh.white:edgecolor=blue \
OAS2_0002_MR2.long.OAS2_0002/surf/rh.white:edgecolor=blueSample Slice 89 in brain.finalsurfs.manedit.mgz, coordinates: 124 138 89 (right hemi)
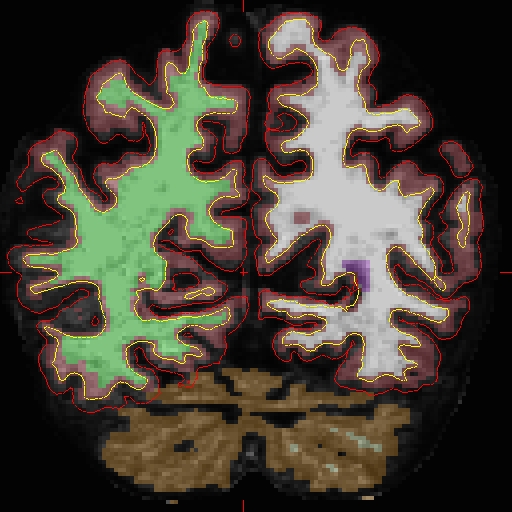
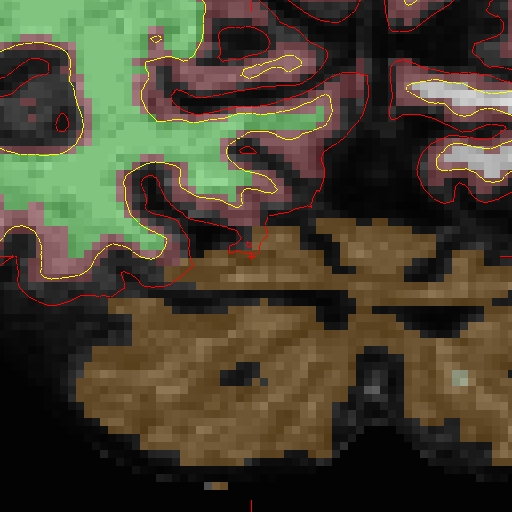
Similarly, create your copy of 'brain.finalsurfs.mgz' with the following command:
cp OAS2_0002_MR1.long.OAS2_0002/mri/brain.finalsurfs.mgz OAS2_0002_MR1.long.OAS2_0002/mri/brain.finalsurfs.manedit.mgz
View the original longitudinal file for MR1 as follows:
freeview -v OAS2_0002_MR1.long.OAS2_0002/mri/brain.finalsurfs.manedit.mgz \
OAS2_0002_MR1.long.OAS2_0002/mri/brainmask.mgz \
OAS2_0002_MR1.long.OAS2_0002/mri/aseg.mgz:colormap=lut:opacity=0.25 \
-f OAS2_0002_MR1.long.OAS2_0002/surf/lh.pial:edgecolor=red \
OAS2_0002_MR1.long.OAS2_0002/surf/rh.pial:edgecolor=red \
OAS2_0002_MR1.long.OAS2_0002/surf/lh.white:edgecolor=blue \
OAS2_0002_MR1.long.OAS2_0002/surf/rh.white:edgecolor=blueAfter edits to both cross-sectional time points and the base have been done, the following commands should be run to re-create new long data sets: (Start with renaming the existing long data to something like _orig) For MR2, you would do:
mv OAS2_0002_MR2.long.OAS2_0002 OAS2_0002_MR2.long.OAS2_0002_orig recon-all -long OAS2_0002_MR2 OAS2_0002 -all
For MR1, you would do:
mv OAS2_0002_MR1.long.OAS2_0002 OAS2_0002_MR1.long.OAS2_0002_orig recon-all -long OAS2_0002_MR1 OAS2_0002 -all
In the re-run the edits are automatically transferred from the cross-sectional. Also the fixed surfaces will be used from the base for initialization producing fixed results in the long run. The results of long re-run can be checked as follows for MR2:
freeview -v OAS2_0002_MR2.long.OAS2_0002_fixed/mri/brain.finalsurfs.mgz \
OAS2_0002_MR2.long.OAS2_0002_fixed/mri/brainmask.mgz \
OAS2_0002_MR2.long.OAS2_0002_fixed/mri/aseg.mgz:colormap=lut:opacity=0.25 \
-f OAS2_0002_MR2.long.OAS2_0002_fixed/surf/lh.pial:edgecolor=red \
OAS2_0002_MR2.long.OAS2_0002_fixed/surf/rh.pial:edgecolor=red \
OAS2_0002_MR2.long.OAS2_0002_fixed/surf/lh.white:edgecolor=blue \
OAS2_0002_MR2.long.OAS2_0002_fixed/surf/rh.white:edgecolor=blueThe following shows pictures of sample slice 89 from OAS2_0002_MR2.long.OAS2_0002_fixed in the brain.finalsurfs.mgz volume at coordinates 112 166 89 :
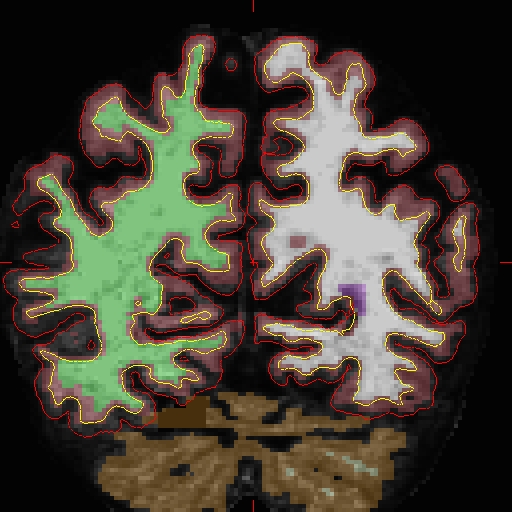
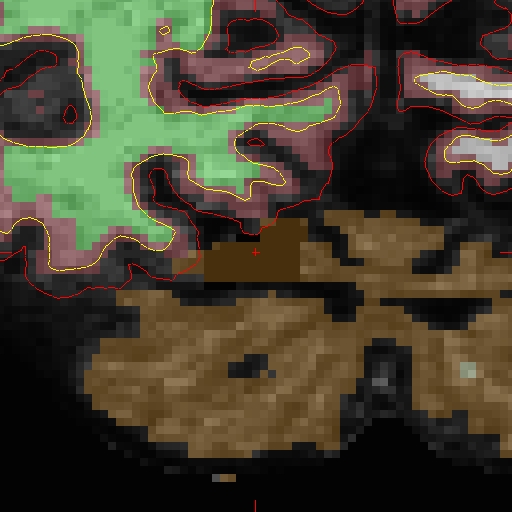
For MR1, look at the results of re-running here:
freeview -v OAS2_0002_MR1.long.OAS2_0002_fixed/mri/brain.finalsurfs.mgz \
OAS2_0002_MR1.long.OAS2_0002_fixed/mri/brainmask.mgz \
OAS2_0002_MR1.long.OAS2_0002_fixed/mri/aseg.mgz:colormap=lut:opacity=0.25 \
-f OAS2_0002_MR1.long.OAS2_0002_fixed/surf/lh.pial:edgecolor=red \
OAS2_0002_MR1.long.OAS2_0002_fixed/surf/rh.pial:edgecolor=red \
OAS2_0002_MR1.long.OAS2_0002_fixed/surf/lh.white:edgecolor=blue \
OAS2_0002_MR1.long.OAS2_0002_fixed/surf/rh.white:edgecolor=blue