|
Size: 6386
Comment:
|
← Revision 28 as of 2013-11-01 14:28:54 ⇥
Size: 7162
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| [wiki:Self:FsTutorial top] | [wiki:Self:FsTutorial/MorphAndRecon previous] | ## page was renamed from FsTutorial/Talairach [[FsTutorial|top]] | [[FsTutorial/TroubleshootingData|previous]] = Fixing Bad Output From the Talairach Registration = *To follow this exercise exactly be sure you've downloaded the [[FsTutorial/Data|tutorial data set]] before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names. |
| Line 3: | Line 6: |
| == Fixing Bad Output From the Talairach Registration == FreeSurfer uses some of the tools developed at the Montreal Neurological Institute (MNI) (http://www.bic.mni.mcgill.ca) to align anatomical volumes (using minctracc) and to compute the linear Talairach transform (using mritotal). The mritotal documentation says that it generally works well for T1 dominant images. The transform is contained in a 3x4 matrix in a file called talairach.xfm and is located in the <subject name>/mri/transforms directory. The mritotal program uses correlation as a similarity measure, and can fail due to image artifacts that reduce the correlation of a good match. Some examples of these are caused by the volume having systematic gray scale variations, noisy white matter regions, or a bright neck region. |
FreeSurfer computes a linear Talairach transform contained in a 3x4 matrix in a file called talairach.xfm and is located in the <subject name>/mri/transforms directory. Under some circumstances, the alignment can fail the automatic failure detection scheme (-tal-check flag, which can be disabled via -notal-check). |
| Line 11: | Line 10: |
| In case of a bad transform, there are some [wiki:Self:FsTutorial/TalairachAuto automatic ways to regenerate the transform]. If the transform still looks bad after trying the automatic methods, you can try a manual registration described below. | Subject tal_before is an example of a bad talairach. The surface cutting planes are far off center but the problem actually originates from the bad talairach transform. To view this talairach in tkmedit first be sure you have the subject open: ---- {{{ tkmedit tal_before brainmask.mgz }}} ---- If the surfaces are also loaded into tkmedit you may find it easier if you toggle them off for this part. |
| Line 13: | Line 18: |
| Go into the talairach_tutorial directory, set your SUBJECTS_DIR environment variable to the current directory, and open the 095_before brain volume with tkmedit: | In the tkmedit toolbar, go to '''File -> Transforms -> Load Transform for Main Volume...''' Click browse, select the file talairach.xfm and click 'OK'. In the coronal view, the transform has resulted in an extremely distorted brain. |
| Line 15: | Line 20: |
| {{attachment:tal_before.jpg}} To further view and edit this transform we will use tkregister2. To open this you can close the tkmedit window and open tkregister2 with the command: ---- |
|
| Line 16: | Line 25: |
| cd $FREESURFER_HOME/subjects/buckner_data/tutorial_subjs setenv SUBJECTS_DIR ${PWD} tkmedit 095_talairach_before brain.mgz & |
tkregister2 --mgz --s tal_before --fstal --surf orig |
| Line 20: | Line 27: |
In the tkmedit toolbar, go to: '''File -> Transforms -> Load Transform for Main Volume...''' Click browse, select the file talairach.xfm and click 'OK'. As you can see in the coronal view, the transform has resulted in an extremely distorted brain. Now, run tkregister2 to open an interface to adjust the registration: {{{ tkregister2 --mgz --s 095_talairach_before --fstal --surf orig }}} |
---- '''Note: If your subject surfaces are not yet available, then exclude the {{{--surf orig}}} flag from tkregister2.''' |
| Line 39: | Line 34: |
| The goal is to stretch, translate, and rotate your MOVEABLE volume so that the two brains look as similar as possible, at least along the key anatomical points (anterior/posterior commissures, the temporal lobes in the coronal plane, and the midline cut). | The goal is to stretch, translate, and rotate your MOVEABLE volume so that the two brains look as similar as possible, at least along the key anatomical points (anterior/posterior commissures, the temporal lobes in the coronal plane, and the midline cut). |
| Line 41: | Line 36: |
| Use Ctrl-1 and Ctrl-2 to switch between the two volumes (or hit the COMPARE button). You will want to do this frequently to check your progress. Click the SAGITTAL button to switch to a sagittal view, and go to slice 128 by using the slider directly below the SAGITTAL button. In this view you have a good view of the corpus callosum in both the moveable and target volumes. | Use Ctrl-1 and Ctrl-2 to switch between the two volumes (or hit the COMPARE button). You will want to do this frequently to check your progress. Click the SAGITTAL button to switch to a sagittal view, and go to slice 128 by using the slider directly below the SAGITTAL button. In this view you have a good view of the corpus callosum in both the moveable and target volumes. |
| Line 43: | Line 38: |
| attachment:tal0.jpg | {{attachment:tal.jpg}} |
| Line 45: | Line 40: |
| To rotate the moveable volume, use the ROTATE BRAIN slider. You will want to move the slider only a couple of degrees at a time until you achieve the desired effect. You will notice a small red cross icon near the middle of the viewing window. This is the center of rotation. You can change the location of the center of rotation by left-clicking in the viewing window with your mouse. | To rotate the moveable volume, use the ROTATE BRAIN slider. You will want to move the slider only a couple of degrees at a time until you achieve the desired effect. You will notice a small red cross icon near the middle of the viewing window. This is the center of rotation. You can change the location of the center of rotation by left-clicking in the viewing window with your mouse. |
| Line 47: | Line 42: |
| attachment:tal1.jpg | {{attachment:tal_before2.jpg}} |
| Line 51: | Line 46: |
| attachment:tal2.jpg | {{attachment:tal_before3.jpg}} |
| Line 53: | Line 48: |
| Once you have the corpus callosum aligned as well as possible in the sagittal plane, click the HORIZONTAL button to get a horizontal view, and use the slider directly below the HORIZONTAL button to go to slice 128. Use Ctrl-1 and Ctrl-2 to switch between the two volumes. Use the ROTATE BRAIN and TRANSLATE BRAIN buttons as before to align the midlines of both volumes. | Once you have the corpus callosum aligned as well as possible in the sagittal plane, click the HORIZONTAL button to get a horizontal view, and use the slider directly below the HORIZONTAL button to go to slice 128. Use Ctrl-1 and Ctrl-2 to switch between the two volumes. Use the ROTATE BRAIN and TRANSLATE BRAIN buttons as before to align the midlines of both volumes. |
| Line 55: | Line 50: |
| attachment:tal3.jpg | {{attachment:tal_before5.jpg}} |
| Line 59: | Line 54: |
| attachment:tal4.jpg | {{attachment:tal_before6.jpg}} |
| Line 63: | Line 58: |
| attachment:tal5.jpg | {{attachment:tal_before7.jpg}} |
| Line 65: | Line 60: |
| Continue this way, switching frequently between the HORIZONTAL, SAGITTAL, and CORONAL views, and align the visible brain structures as much as possible in all of the slices. Use the SCALE BRAIN button as needed to scale the brain in the X and Y direction. Keep in mind that you are working in 3D, not in 2D, so any changes made in one view will affect the other views as well. attachment:talcor.jpg attachment:talsag.jpg attachment:talhor.jpg |
Continue this way, switching frequently between the HORIZONTAL, SAGITTAL, and CORONAL views, and align the visible brain structures as much as possible in all of the slices. Use the SCALE BRAIN button as needed to scale the brain in the X and Y direction. Keep in mind that you are working in 3D, not in 2D, so any changes made in one view will affect the other views as well. |
| Line 71: | Line 62: |
| When you are satisfied with your registration result, click the SAVE REG button in the tkregister2 toolbar. | {{attachment:tal_before8.jpg}} {{attachment:talbefore9.jpg}} {{attachment:talbefore10.jpg}} When you are satisfied with your registration result, click the SAVE REG button in the tkregister2 toolbar. |
| Line 74: | Line 69: |
| '''Note: do not run the Talairach stage of the FreeSurfer reconstruction or your manual edits will be overwritten. E.g., when running recon-all, use the -notalairach flag.''' | Because the talairach transformation affects everything in the stream it is necessary to rerun the whole process using your new talairach, which is a default. You can do this with the command: {{{ recon-all -all -subjid tal_before }}} This step will take a long time to run, it is not necessary to run this step for the purposes of this tutorial. You can compare your new talairach to the one in tal_after. |
| Line 76: | Line 75: |
| Lastly, there is an alternate means of producing a talairach transform that may work better in some cases. To run that method: {{{ recon-all -s tal_before -talairach -use-mritotal -tal-check -clean-tal }}} This runs the {{{mritotal}}} utility from the MNI toolset. Note that for this particular subject example, it happens to produce worse results than the default talairach transform method. Also note that the -clean-tal flag will delete any existing talairach.xfm file. See also [[TalFailV5.1]]. |
Fixing Bad Output From the Talairach Registration
To follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names.
FreeSurfer computes a linear Talairach transform contained in a 3x4 matrix in a file called talairach.xfm and is located in the <subject name>/mri/transforms directory. Under some circumstances, the alignment can fail the automatic failure detection scheme (-tal-check flag, which can be disabled via -notal-check).
The best way to check the transform is by loading it visually. This can be done directly from tkmedit (File --> Load Transform), however new users sometimes feel more comfortable viewing the transform in tkregister2, where it is possible to view the transform on top of the talairach subject. When viewing in tkmedit you can expect some distortion and stretching from the position of your subject - especially a sagittal tilt, but you want to avoid any changes in orientation (i.e., the coronal view of your subject should be the coronal view of your talairach transform), severe changes in positioning in the window (i.e., your transformation should not show up in the top left corner of the view, while your subject is nicely centered), and rotations or twisting. If any of these are seen, you will need to correct your transform.
Subject tal_before is an example of a bad talairach. The surface cutting planes are far off center but the problem actually originates from the bad talairach transform. To view this talairach in tkmedit first be sure you have the subject open:
tkmedit tal_before brainmask.mgz
If the surfaces are also loaded into tkmedit you may find it easier if you toggle them off for this part.
In the tkmedit toolbar, go to File -> Transforms -> Load Transform for Main Volume... Click browse, select the file talairach.xfm and click 'OK'. In the coronal view, the transform has resulted in an extremely distorted brain.
To further view and edit this transform we will use tkregister2. To open this you can close the tkmedit window and open tkregister2 with the command:
tkregister2 --mgz --s tal_before --fstal --surf orig
Note: If your subject surfaces are not yet available, then exclude the --surf orig flag from tkregister2.
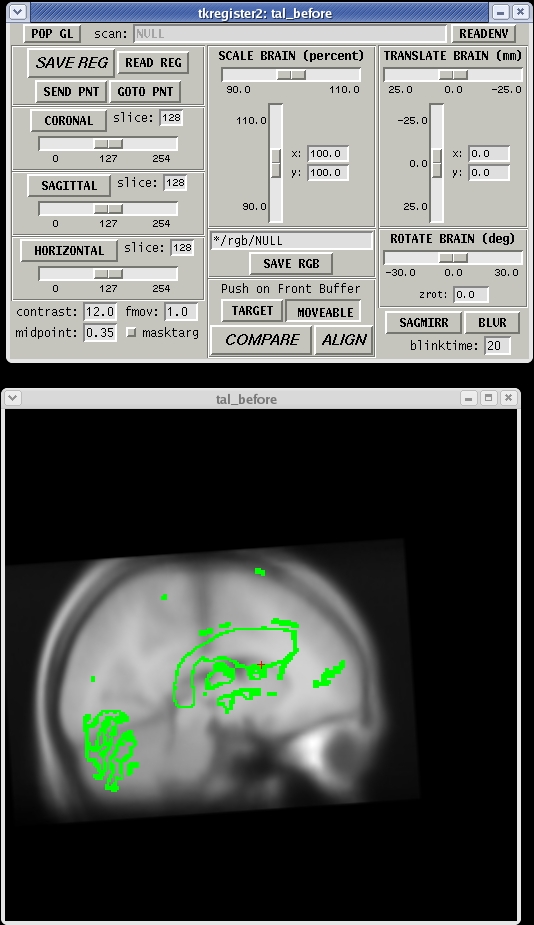
Brief descriptions on how to operate tkregister2 are given below. More documentation is also available by running "tkregister2 --help".
You will see the subject's volume as the TARGET volume and your Talairach volume as a fuzzy MOVEABLE volume. The green lines are the orig surface from the subject. This will be the same in both the TARGET and the MOVEABLE. It can be turned on and off by clicking in the image window and hitting the 's' key.. Find the fmov: box in the tkregister toolbar and make sure it is set to 1.0.
The goal is to stretch, translate, and rotate your MOVEABLE volume so that the two brains look as similar as possible, at least along the key anatomical points (anterior/posterior commissures, the temporal lobes in the coronal plane, and the midline cut).
Use Ctrl-1 and Ctrl-2 to switch between the two volumes (or hit the COMPARE button). You will want to do this frequently to check your progress. Click the SAGITTAL button to switch to a sagittal view, and go to slice 128 by using the slider directly below the SAGITTAL button. In this view you have a good view of the corpus callosum in both the moveable and target volumes.
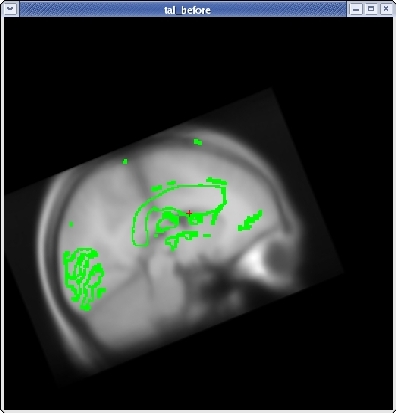
To rotate the moveable volume, use the ROTATE BRAIN slider. You will want to move the slider only a couple of degrees at a time until you achieve the desired effect. You will notice a small red cross icon near the middle of the viewing window. This is the center of rotation. You can change the location of the center of rotation by left-clicking in the viewing window with your mouse.
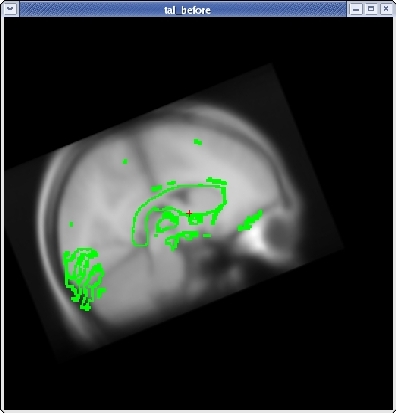
For translation, there are two sliders: one to move the volume left and right, and one to move the volume up and down. Next, translate the moveable brain upwards by using the TRANSLATE BRAIN vertical slider. Again, you only want to move the volume a couple millimeters at a time. You can move the volume left and right in the same way.
Once you have the corpus callosum aligned as well as possible in the sagittal plane, click the HORIZONTAL button to get a horizontal view, and use the slider directly below the HORIZONTAL button to go to slice 128. Use Ctrl-1 and Ctrl-2 to switch between the two volumes. Use the ROTATE BRAIN and TRANSLATE BRAIN buttons as before to align the midlines of both volumes.
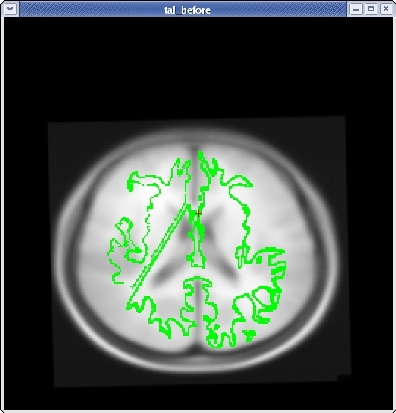
Once you are done aligning the brains in the horizontal view, switch back to slice 128 in the sagittal view. Fine tune your rotation and translation again until the corpus callosum is once again aligned in both volumes.
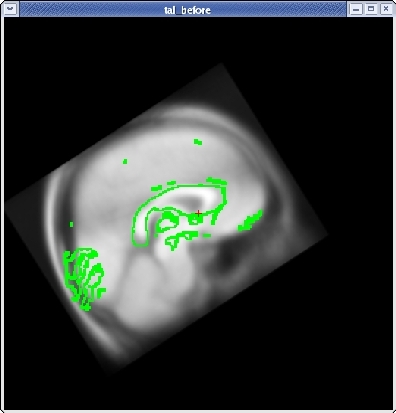
Click the CORONAL button, and go to slice 128. Align the midlines of the brains again in the same way.
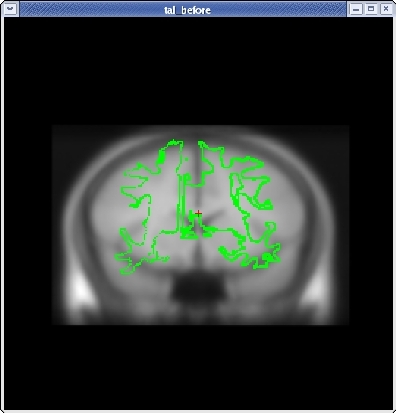
Continue this way, switching frequently between the HORIZONTAL, SAGITTAL, and CORONAL views, and align the visible brain structures as much as possible in all of the slices. Use the SCALE BRAIN button as needed to scale the brain in the X and Y direction. Keep in mind that you are working in 3D, not in 2D, so any changes made in one view will affect the other views as well.
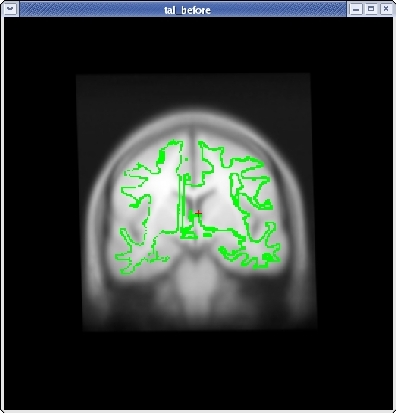
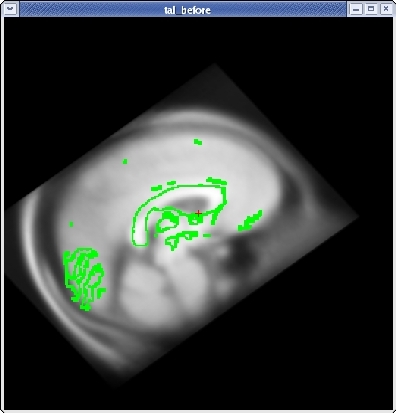
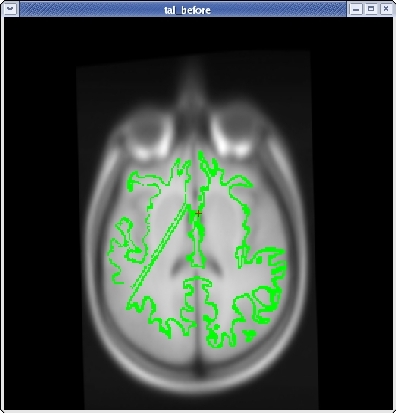
When you are satisfied with your registration result, click the SAVE REG button in the tkregister2 toolbar. You can close tkregister2 by clicking on the red close button in the upper left hand corner of the viewing window, or by typing Ctrl-C in in the same terminal window as the new matrix.
Because the talairach transformation affects everything in the stream it is necessary to rerun the whole process using your new talairach, which is a default. You can do this with the command:
recon-all -all -subjid tal_before
This step will take a long time to run, it is not necessary to run this step for the purposes of this tutorial. You can compare your new talairach to the one in tal_after.
Lastly, there is an alternate means of producing a talairach transform that may work better in some cases. To run that method:
recon-all -s tal_before -talairach -use-mritotal -tal-check -clean-tal
This runs the mritotal utility from the MNI toolset. Note that for this particular subject example, it happens to produce worse results than the default talairach transform method. Also note that the -clean-tal flag will delete any existing talairach.xfm file. See also TalFailV5.1.
