Making white matter edits
To follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names.
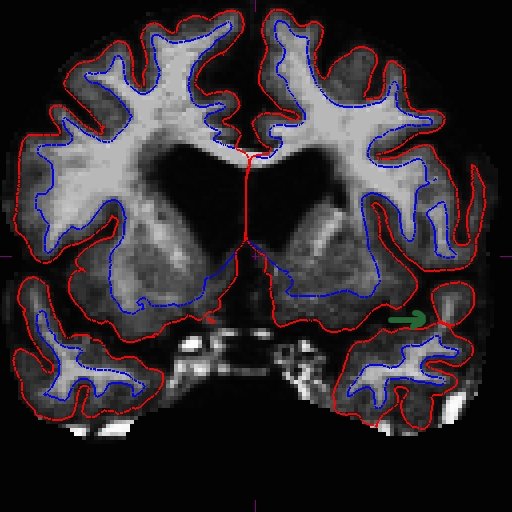
For the longitudinal stream, it is recommended that you make wm.mgz edits to the base. This is because even though the long transfers manual wm edits from the cross, it still takes the surfaces from the base, and if the base surfaces did not improve, neither will the long. The only way to make corrections to the base surfaces themselves is to edit wm.mgz on the base. Therefore, making wm.mgz right on the base can save you a lot of time. Of course it is best to edit all stages (all cross time points and base), but often it is sufficient to edit only the base. Were you able to find the area that needs intervention? Here is what we found. You can see below on the brainmask.mgz (left) that the pial surface is cutting through the wm. If you toggle to the wm.mgz (right), you can see that there are a few wm voxels missing.
This happens at the tip of the lh temporal lobe several slices away (coronal slice 166).
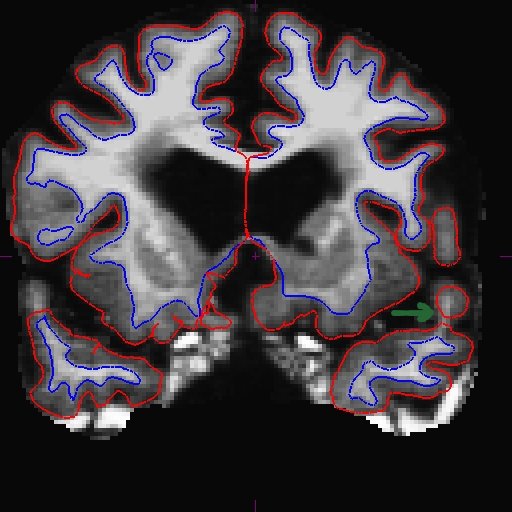
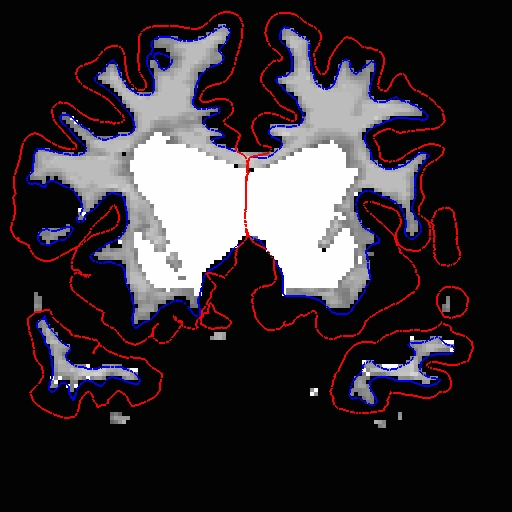
Add wm voxels to connect the wm together in all the slices that need it. Here is what we did.
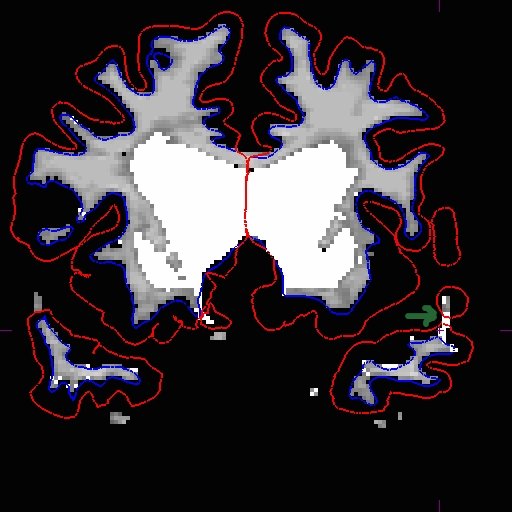
After you are satisfied with your edits, save the aux volume and rerun the base.
Don't run the recon-all commands on this page
It will take hours and has already been done for you.
recon-all -base OAS2_0185 -autorecon2-wm -autorecon3
After that is done, check the surfaces on base using tkmedit.
freeview -v OAS2_0185_fixed/mri/wm.mgz \
OAS2_0185_fixed/mri/brainmask.mgz \
-f OAS2_0185_fixed/surf/lh.pial:edgecolor=red \
OAS2_0185_fixed/surf/rh.pial:edgecolor=red \
OAS2_0185_fixed/surf/lh.white:edgecolor=blue \
OAS2_0185_fixed/surf/rh.white:edgecolor=blueThis is what it should look like now in slice 166.
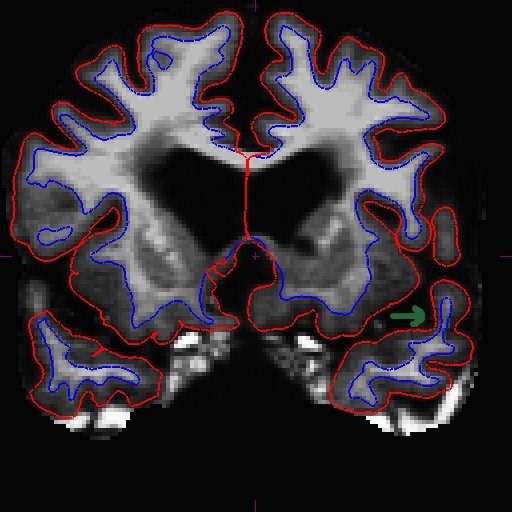
When the surfaces are all corrected, you would recreate the long (make sure you remove the long directories before the rerun - or at the very least, remove the wm.mgz files in all the mri sub-directories of the long runs).
recon-all -long OAS2_0185_MR1 OAS2_0185 -all
recon-all -long OAS2_0185_MR2 OAS2_0185 -all
Inspect your output:
freeview -v OAS2_0185_MR1.long.OAS2_0185_fixed/mri/wm.mgz \
OAS2_0185_MR1.long.OAS2_0185_fixed/mri/brainmask.mgz \
-f OAS2_0185_MR1.long.OAS2_0185_fixed/surf/lh.pial:edgecolor=red \
OAS2_0185_MR1.long.OAS2_0185_fixed/surf/rh.pial:edgecolor=red \
OAS2_0185_MR1.long.OAS2_0185_fixed/surf/lh.white:edgecolor=blue \
OAS2_0185_MR1.long.OAS2_0185_fixed/surf/rh.white:edgecolor=bluefreeview -v OAS2_0185_MR2.long.OAS2_0185_fixed/mri/wm.mgz \
OAS2_0185_MR2.long.OAS2_0185_fixed/mri/brainmask.mgz \
-f OAS2_0185_MR2.long.OAS2_0185_fixed/surf/lh.pial:edgecolor=red \
OAS2_0185_MR2.long.OAS2_0185_fixed/surf/rh.pial:edgecolor=red \
OAS2_0185_MR2.long.OAS2_0185_fixed/surf/lh.white:edgecolor=blue \
OAS2_0185_MR2.long.OAS2_0185_fixed/surf/rh.white:edgecolor=blueSee here that the improved surfaces in the base also correct the surfaces in the long. Compare MR1.long slice 163 in the before (left) and after (right) images below.
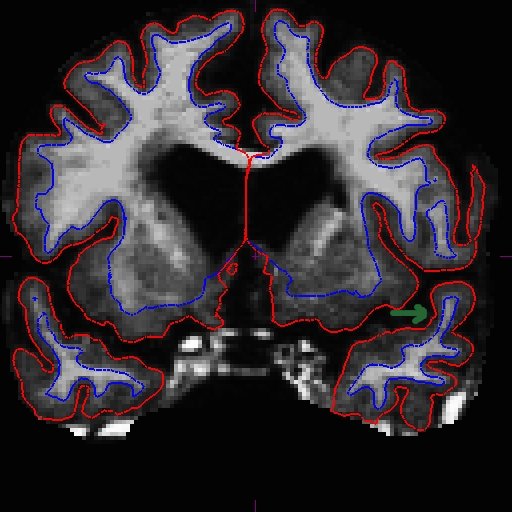
The same improvement should have occurred with MR2.long.
