| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| Line 6: | Line 5: |
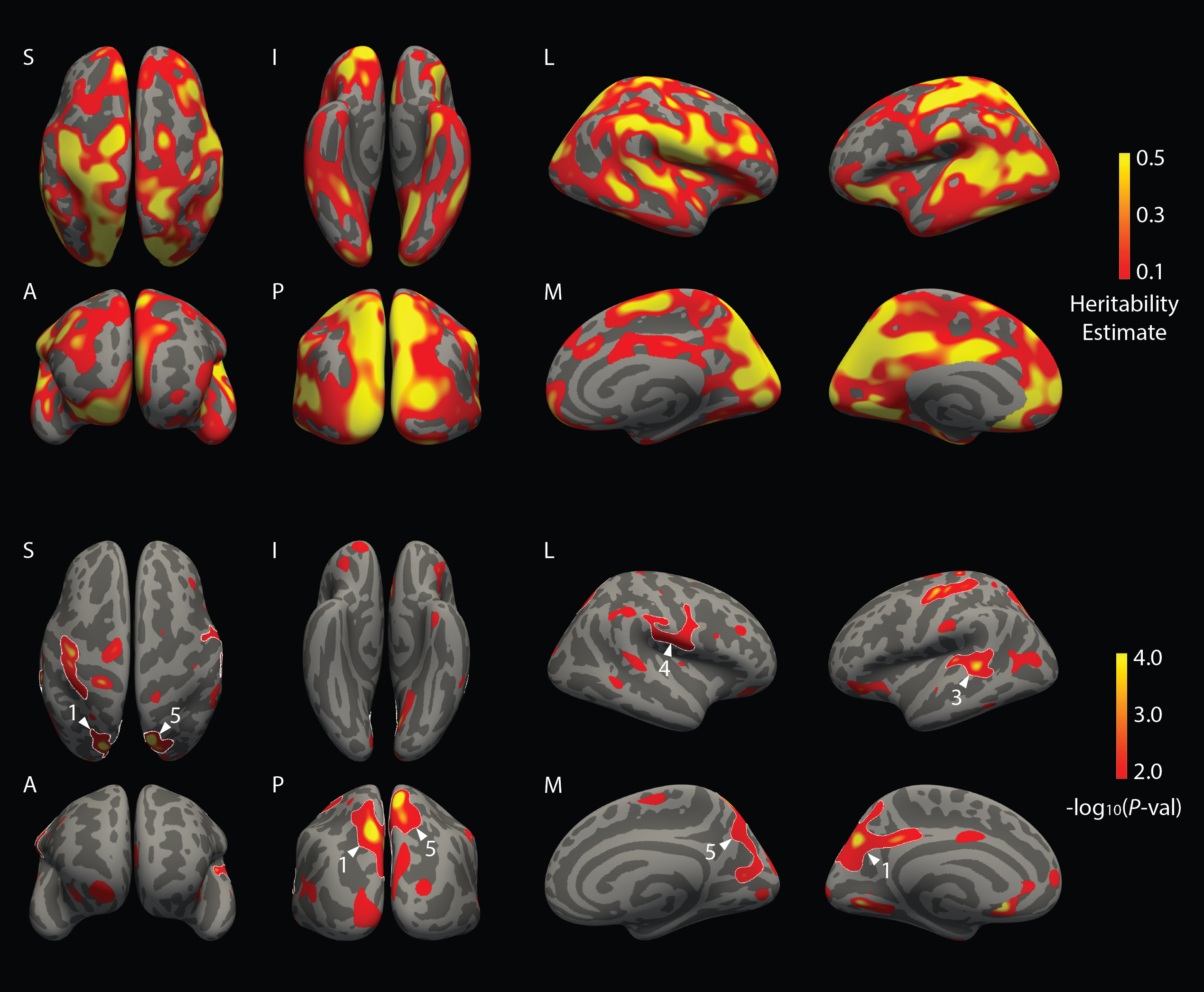
| || We are distributing vertex-wise surface maps of the heritability of cortical thickness, sulcal depth, curvature, and surface area. These maps are in standard FreeSurfer surface space (fsaverage) and can be used as a prior in genetic association studies of cortical surface morphology. For example, researchers can prioritize different regions or morphometric measurements based on the heritability estimates we provide. Alternatively, researchers can use these maps to define regions of interest (ROIs) to focus their analyses on. | |
| Line 7: | Line 7: |
|
|| We are distributing vertex-wise surface maps of the heritability of cortical thickness, sulcal depth, curvature, and surface area. These maps are in standard FreeSurfer surface space (fsaverage) and can be used as a prior in genetic association studies of cortical surface morphology. For example, researchers can prioritize different regions or morphometric measurements based on the heritability estimates we provide. Alternatively, researchers can use these maps to define regions of interest (ROIs) to focus their analyses on. |
These maps were computed using MEGHA (Massively Expedited Genome-wide Heritability Analysis). We developed MEGHA, a fast and accurate statistical method for high-dimensional heritability analysis using genome-wide single nucleotide polymorphism (SNP) data from unrelated individuals, and accompanying nonparametric sampling techniques that enable flexible inferences for arbitrary statistics of interest. MEGHA produces estimates and significance measures of heritability with several orders of magnitude less computational time than existing methods, making heritability-based prioritization of millions of phenotypes based on data from unrelated individuals tractable for the first time. We computed vertex-wise surface maps in FreeSurfer fsaverage space for SNP-based heritability estimates and significance of cortical thickness, sulcal depth, curvature, and surface area measures, using data from 1,320 unrelated young (18-35y) healthy adults of non-Hispanic European ancestry as part of the Harvard/MGH Brain Genomics Superstruct Project (GSP). These surface maps are available for download. |
| Line 11: | Line 9: |
|
These maps were computed using MEGHA (Massively Expedited Genome-wide Heritability Analysis). We developed MEGHA, a fast and accurate statistical method for high-dimensional heritability analysis using genome-wide single nucleotide polymorphism (SNP) data from unrelated individuals, and accompanying nonparametric sampling techniques that enable flexible inferences for arbitrary statistics of interest. MEGHA produces estimates and significance measures of heritability with several orders of magnitude less computational time than existing methods, making heritability-based prioritization of millions of phenotypes based on data from unrelated individuals tractable for the first time. We computed vertex-wise surface maps in FreeSurfer fsaverage space for SNP-based heritability estimates and significance of cortical thickness, sulcal depth, curvature, and surface area measures, using data from 1,320 unrelated young (18-35y) healthy adults of non-Hispanic European ancestry as part of the Harvard/MGH Brain Genomics Superstruct Project (GSP). These surface maps are available for download. || {{attachment:MEGHA_Ge2015.png | Heritability | width=300 height=180}} || |
|| {{attachment:MEGHA_Ge2015.png|Heritability|width="300 height=180"}} || |
| Line 15: | Line 12: |
|
[[http://scholar.harvard.edu/files/tge/files/massively_expedited_genome-wide_heritability_analysis_megha.pdf | Ge T, Nichols TE, Lee PH, Holmes AJ, Roffman JL, Buckner RL, Sabuncu MR*, Smoller JW*. Massively Expedited Genome-wide Heritability Analysis (MEGHA), Proceedings of the National Academy of Sciences, 112(8), 2479-2484. (*contributed equally)]] |
[[http://scholar.harvard.edu/files/tge/files/massively_expedited_genome-wide_heritability_analysis_megha.pdf|Ge T, Nichols TE, Lee PH, Holmes AJ, Roffman JL, Buckner RL, Sabuncu MR*, Smoller JW*. Massively Expedited Genome-wide Heritability Analysis (MEGHA), Proceedings of the National Academy of Sciences, 112(8), 2479-2484. (*contributed equally)]] |
| Line 19: | Line 15: |
|
A MATLAB implementation of MEGHA can be found [[http://scholar.harvard.edu/tge/software/megha | here]]. |
A MATLAB implementation of MEGHA can be found [[http://scholar.harvard.edu/tge/software/megha|here]]. |
| Line 23: | Line 18: |
|
{{attachment:SurfaceMap_Ge2015.png | Surface Maps | height=500}} |
{{attachment:SurfaceMap_Ge2015.png|Surface Maps|height="500"}} |
| Line 27: | Line 21: |
|
[[attachment:SurfMaps.zip | Vertex-wise surface maps for SNP-based heritability estimates and significance of cortical thickness, sulcal depth, curvature, and surface area measures]]. These maps will be included in future FreeSurfer releases. |
[[attachment:SurfMaps.zip|Vertex-wise surface maps for SNP-based heritability estimates and significance of cortical thickness, sulcal depth, curvature, and surface area measures]]. These maps will be included in future FreeSurfer releases. |
Heritability Maps of Cortical Structure Estimated with MEGHA
|| We are distributing vertex-wise surface maps of the heritability of cortical thickness, sulcal depth, curvature, and surface area. These maps are in standard FreeSurfer surface space (fsaverage) and can be used as a prior in genetic association studies of cortical surface morphology. For example, researchers can prioritize different regions or morphometric measurements based on the heritability estimates we provide. Alternatively, researchers can use these maps to define regions of interest (ROIs) to focus their analyses on.
These maps were computed using MEGHA (Massively Expedited Genome-wide Heritability Analysis). We developed MEGHA, a fast and accurate statistical method for high-dimensional heritability analysis using genome-wide single nucleotide polymorphism (SNP) data from unrelated individuals, and accompanying nonparametric sampling techniques that enable flexible inferences for arbitrary statistics of interest. MEGHA produces estimates and significance measures of heritability with several orders of magnitude less computational time than existing methods, making heritability-based prioritization of millions of phenotypes based on data from unrelated individuals tractable for the first time. We computed vertex-wise surface maps in FreeSurfer fsaverage space for SNP-based heritability estimates and significance of cortical thickness, sulcal depth, curvature, and surface area measures, using data from 1,320 unrelated young (18-35y) healthy adults of non-Hispanic European ancestry as part of the Harvard/MGH Brain Genomics Superstruct Project (GSP). These surface maps are available for download.
|
References
Software
A MATLAB implementation of MEGHA can be found here.
Surface Maps for Cortical Thickness Measures
Downloads
Vertex-wise surface maps for SNP-based heritability estimates and significance of cortical thickness, sulcal depth, curvature, and surface area measures. These maps will be included in future FreeSurfer releases.

