| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| [[attachment:data_checker.tgz]] |
STATUS
scripts are all here: /autofs/space/tensor_017/users/jpacheco/QAtools * they should all be working now. To-Do: *(standard_in) 1: parse error * check group stats scripts. Things I've done: * I've created the pages ReconChecker and QAnotepad which will be the only place that full usage options for these scripts will be, eventually. * VP added 3 scripts to the data_checker directory: gnicv, gparcvalnorm, gparcmeannorm. Now the recon_all_aseg_outlier_checker now compares and looks for outliers after the segmentation volumes have been corrected for ICV, per David's suggestion * add a flag for -noasegoutlier - DONE by vp on 9/28/06 * make version checker scripts output to the same log file as recon_checker - DONE by jp on 9/29/06 * figure out a better way to define RECON_CHECKER_SCRIPTS and scriptDirs....INSTEAD, set QA_SCRIPTS to be the QAtools directory, and then within the scripts set RECON_CHECKER_SCRIPTS to be $QA_SCRIPTS/data_checker/ - DONE by jp 10/12/06 * add greconversion and greconversions to ALWAYS run first when the recon-checker is run - DONE by jp on 09/29/06 * -outputfileorder appears to not work. - DONE by vp 10/10/06 * i think i can fix the snapshot script to make it run faster. - DONE by jp 10/12/06 * figure out how to make an aseg look up table. I did this, added the flag -makeAsegLookUp to the recon_checker script. - DONE by jp 11/09/06
FreeSurfer QA Tools
Contents
QA mini-intro coming soon...
To check the output of one reconned subject
First you'll need to properly set your SUBJECTS_DIR. Also you'll need to set your QA_SCRIPTS directory. This should be the directory that you downloaded the QAtools directory. For our example here QAtools is in /space/tensor/17/users/jpacheco/QAtools.
setenv QA_SCRIPTS /space/tensor/17/users/jpacheco/QAtools
Now, from your SUBJECTS_DIR you are ready to run:
$QA_SCRIPTS/data_checker/recon_checker -nocheckasegoutliers -s bert -o QAsnap.html
where bert is your subject ID and QAsnap.html is the name of the output html file that you want to create.
For complete usage instructions please see the ReconChecker page.
What it does:
checks the version of FreeSurfer used to process this subject
- checks the order of the output files to be sure that they were created in the correct order
checks the recon-all-status.log file to be sure each step was run
- takes snapshots of the talairach, skullstrip, surfaces, curvature, and parcellations
- outputs two different log files and an html file (called QAsnap.html in this example) with the snapshots
**NOTE: while it takes the snapshots tkmedit and tksurfer will flash up on the screen. You do not need to do anything, the snap shots are taken automatically. To ensure good quality snapshots make sure there are no other windows over the tkmedit/tksurfer windows.
Outputs
There will be two log files output into your subjects scripts directory:
ls bert/scripts recon_checker.bert.details.log recon_checker.bert.summary.log
The recon_checker.bert.summary.log contains a summary of the status of that subject:
------------------------------------------------------------- Running recon_checker on: Thu Oct 12 14:51:24 EDT 2006 Checking Last Version Used: /usr/local/freesurfer/dev/bin/recon-all In output_file_checker checking if output file order file is set... output_file_order_file not specified, using default order check to see if all files exist... bert outputfiles_OK In recon_all_status_log_checker bert COMPLETE
This subject was run most recently using the dev version of FreeSurfer, all of the output files have been created in the correct order, and all the steps have been run. Another example of this log file is:
------------------------------------------------------------- Running recon_checker on: Thu Oct 12 14:54:24 EDT 2006 Checking Last Version Used: /space/dijon/28/users/jpacheco/freesurfer_stable3_centos64//bin/recon-all In output_file_checker checking if output file order file is set... output_file_order_file not specified, using default order check to see if all files exist... bert outputfiles_NOTOK In recon_all_status_log_checker bert STEPS_MISSING
This subject was run most recently using a version of FreeSurfer located /space/dijon/28/users/jpacheco/freesurfer_stable3_centos64, the output files are not in the ok order, and there are steps missing from the processing stream.
The recon_checker.bert.details.log will give more details as to what might be wrong if there are problems. For the second subject above, with steps missing, the recon_checker.bert.details.log looks like:
------------------------------------------------------------- Running recon_checker on: Thu Oct 12 14:54:24 EDT 2006 Checking Last Version Used: /space/dijon/28/users/jpacheco/freesurfer_stable3_centos64//bin/recon-all Checking ALL Versions Used: /usr/local/freesurfer/dev/bin/recon-all, /space/dijon/28/users/jpacheco/freesurfer_stable3_centos64//bin/recon-all, In output_file_checker SUBJECTS_DIR is /autofs/space/tensor_014/users/jenni checking if output file order file is set... output_file_order_file not specified, using default order realOrder_plus1 is 72 /autofs/space/tensor_014/users/bert/mri/orig/001.mgz last modified 2005-11-02 12:53:09.000000000 -0500 /autofs/space/tensor_014/users/bert/mri/orig/002.mgz last modified 2005-11-02 12:53:56.000000000 -0500 ERROR: order is not commmon order: stopped at mri/transforms/talairach.xfm, should be mri/rawavg.mgz ... real order: common order: .......... ......... .......... ......... .......... ......... /autofs/space/tensor_014/users/bert/mri/orig/001.mgz /autofs/space/tensor_014/users/bert/mri/orig/001.mgz /autofs/space/tensor_014/users/bert/mri/orig/002.mgz /autofs/space/tensor_014/users/bert/mri/orig/002.mgz mri/transforms/talairach.xfm mri/rawavg.mgz mri/rawavg.mgz mri/orig.mgz mri/orig.mgz mri/nu.mgz mri/nu.mgz mri/transforms/talairach.auto.xfm mri/transforms/talairach.auto.xfm mri/transforms/talairach.xfm .......... ......... .......... ......... .......... ......... bert outputfiles_NOTOK In recon_all_status_log_checker bert STEPS_MISSING Expected to see the following steps before #@# Tessellate rh Wed Jun 7 14:43:02 EDT 2006 in the recon-all-status.log file (line # 98): --> Tessellate lh --> Smooth1 lh --> Inflation1 lh --> QSphere lh --> Fix Topology lh --> Make Final Surf lh --> Smooth2 lh --> Inflation2 lh --> Cortical ribbon mask lh --> Sphere lh --> Surf Reg lh --> Contra Surf Reg lh --> AvgCurv lh --> Cortical Parc lh --> Parcellation Stats lh --> Cortical Parc 2 lh --> Parcellation Stats 2 lh However, these LH processes were executed previously. Perhaps the RH output was modified and re-run without re-running the LH processes.
**NOTE: a portion of this log file was excluded for simplification of display purposes. Those lines included a list of the full paths to the 71 files being checked.
This log shows that the dev version of FreeSurfer was used for a previous set of processing, but most recently the private version was used. The output from the file order checking shows that the rawavg.mgz file was created after the transforms/talairach.xfm file but it was supposed to be created first. The file order checking was stopped there. The recon-all-status.log checker shows that all the LH processes are out of order. It offers a hint of explanation, saying that the RH may have been re-run without the LH since the LH processes were completed earlier. For this subject that was the case.
Lastly, an html file was created, QAsnap.html, and can be viewed in any standard browser. You can open it from the command line
mozilla $SUBJECTS_DIR/QAsnap.html
or put the path to your file directly in the browser. This will show you a web report of all the subjects you just ran. In our case we've only run one subject so we've only got one subject in our list
When you click on bert snapshots it will take you to a page with all the snapshots from that subject.
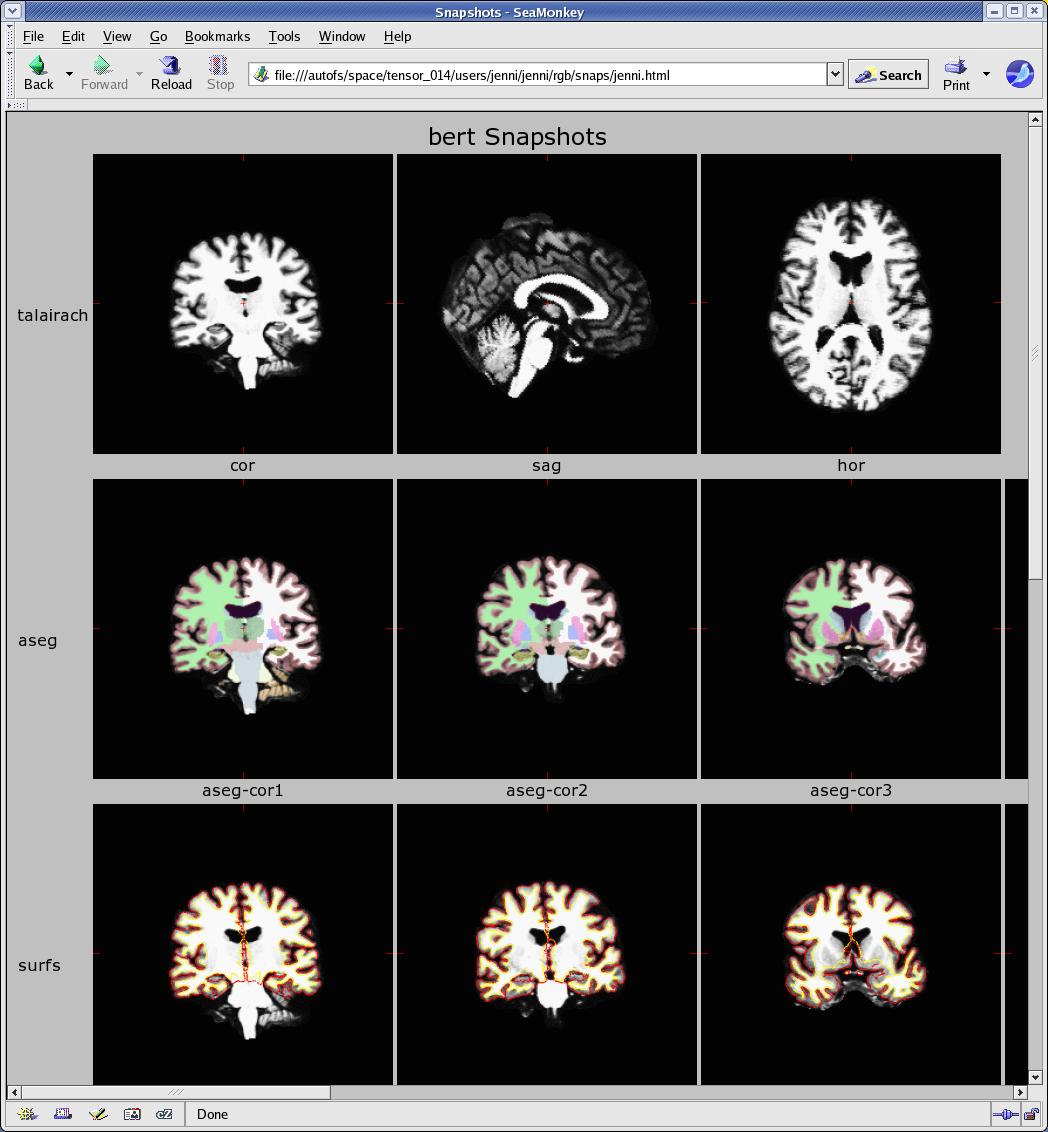
**NOTE: this shows just a portion of the snapshot output page, there are more snapshots if you scroll right and down.
Usage Recommendations
When you go through and check your outputs you can open the QAnotepad to keep track of your observations. A simple comand line to open the QAnotepad is:
$QA_SCRIPTS/QAnotepad -s <subject1> <subject2> ...
or, if you'd like to pass it your own config file then you can use:
$QA_SCRIPTS/QAnotepad -s <subject1> <subject2> ... -c <config file> -f <output file name>
The QAnotepad is a customizable tool that handles notes (both radio-button selections and free text) on various aspects of the FreeSurfer output, for further usage options and explanations, including how to design your own config file, see the QAnotepad wiki page.
To check the output of a group of reconned subjects
First you'll need to properly set your SUBJECTS_DIR. Also you'll need to set your QA_SCRIPTS directory. This should be the directory that you downloaded the QAtools directory. For our example here QAtools is in /space/tensor/17/users/jpacheco/QAtools.
setenv QA_SCRIPTS /space/tensor/17/users/jpacheco/QAtools
The first step is to make a aseg outliers look up table. You can run this command to do that:
$QA_SCRIPTS/data_checker/recon_checker -s <list of subjects> -makeAsegLookUp AsegLookUp.txt
where <list of subjects> is your own list of subjects, from which you want to make a look-up table. This will generate a file named AsegLookUp.txt, or whatever you have specified after the '-makeAsegLookUp' flag.
This command will generate an aseg look-up table, that can be used in subsequent runs. It will also check the versions used for each subject and check for outliers based on the table it has just created. The version and outlier information is saved into the summary and detail logs (see below for more information). It will NOT take snapshots or check any other ouptus, if you want to take snapshots, or check for other options as well you will need to add those additional flags to the command line. (see ReconChecker for more usage and options).
If you have already made your look-up table, but need to check for outliers again you can run the command:
$QA_SCRIPTS/data_checker/recon_checker -s <list of subjects> -checkasegoutliers -asegMeansFile AsegLookUp.txt
where <list of subjects> is your own list of subjects and AsegLookUp.txt is the name of your look-up table.
This command will use a previously made look-up table to check for aseg outliers. It will also check the versions used, the output files, the status log order, and take snapshots. If you want to skip some of these options you will need to add those additional flags to the command line. (see ReconChecker for more usage and options).
Outputs
Usage Recommendations
It's a good idea to make a seperate look-up table for each group in your study (i.e., in a study of patients versus controls make a look-up table for patients and another for controls) To do this you could run these commands:
$QA_SCRIPTS/data_checker/recon_checker -s $controls -makeAsegLookUp AsegLookUp_control.txt $QA_SCRIPTS/data_checker/recon_checker -s $patients -makeAsegLookUp AsegLookUp_patient.txt
Once you have your group identified it's a good idea to rerun the -takesnapshots option. This will regenerate the opening html file, with a listing of all your subjects, making it easier to check the snapshots if needed.
