recon-any
This functionality is available dev version of FreeSurfer. Author: Karthik Gopinath E-mail: kgopinath[at]mgh[dot]harvard[dot]edu Please post your questions on this module to the FreeSurfer mailing list at freesurfer[at]nmr.mgh.harvard.edu rather than directly contacting the author. If you use this package in your analysis, please cite:
Cortical analysis of heterogeneous clinical brain MRI scans for large-scale neuroimaging studies. K Gopinath, DN Greeve, S Das, S Arnold, C Magdamo, JE Iglesias
SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining. B Billot, DN Greve, O Puonti, A Thielscher, K Van Leemput, B Fischl, AV Dalca, JE Iglesias. Medical Image Analysis, 83, 102789 (2023).
Robust machine learning segmentation for large-scale analysis of heterogeneous clinical brain MRI datasets. B Billot, C Magdamo, SE Arnold, S Das, JE Iglesias. PNAS, 120(9), e2216399120 (2023).
SynthSR: a public AI tool to turn heterogeneous clinical brain scans into high-resolution T1-weighted images for 3D morphometry. JE Iglesias, B Billot, Y Balbastre, C Magdamo, S Arnold, S Das, B Edlow, D Alexander, P Golland, B Fischl. Science Advances, 9(5), eadd3607 (2023).
General description:
This tool performs recon-any, the first out-of-the-box cortical surface reconstruction and analysis of brain MRI scans of any modality, contrast and resolution without retraining and fine-tuning. This "Recon-all-like" stream for any adult 3D brain scans is essentially a combination of:
SynthSeg: to obtain a volumetric segmentation and linear registration to Talairach space
SynthSR: to have a higher resolution 1mm MPRAGE for visualization
SynthDist: to fit surfaces by predicting the distance maps and reconstructing topologically accurate cortical surfaces
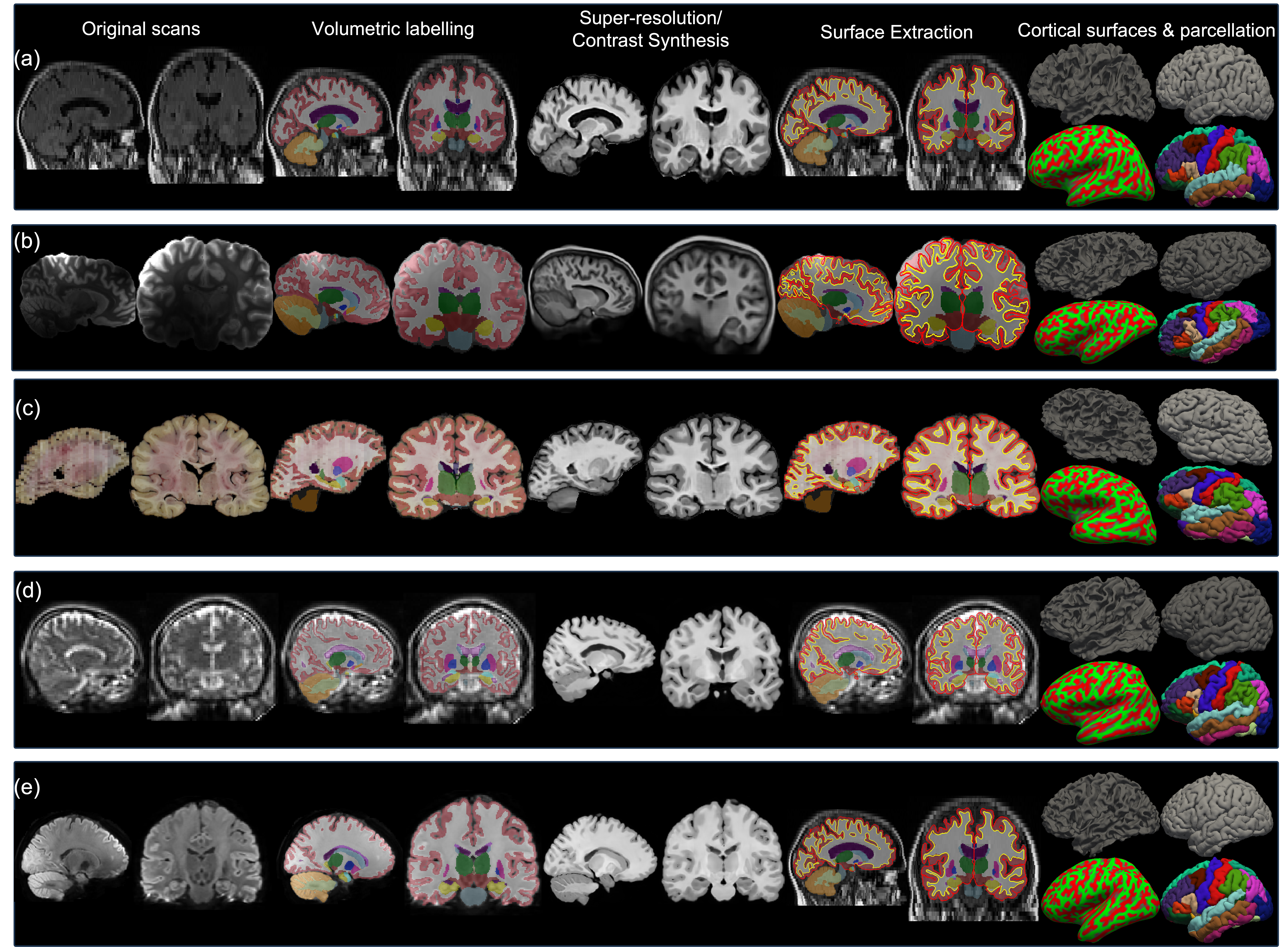
Sample outputs of cortical surfaces for different scans from the different datasets. (a) Axial T1-weighted scan with 2×2×2mm resolution. (b) Ex vivo MRI scan at 0.5×0.5×0.5mm resolution. (c) A 3D reconstruction of the coronal dissection photograph at 0.5×0.5×4mm. (d) A portable low-field T2 axial scan at 2.5x2.5x2.5mm resolution. (e) A diffusion-MRI scan at 1.25×1.25×1.25mm resolution.
Usage:
OnceFreeSurfer has been sourced, you can simply run recon-all-clinical on your own data with
recon-all-clinical.sh INPUT_SCAN SUBJECT_ID THREADS [SUBJECT_DIR]
where: - INPUT_SCAN: path to an image that will be processed. - SUBJECT_ID: specifies the name or ID of the subject you would like to use. A directory with that name will be created for all the subject's FreeSurfer output. - THREADS (optional): number of CPU threads to use. The default is just 1, so crank it up for faster processing if you have multiple cores! - SUBJECT_DIR: only necessary if the environment variable SUBJECTS_DIR has not been set when sourcing FreeSurfer or if you want to override it. This stream runs a bit faster than the original recon-all, since the volumetric segmentation is much faster than the iterative Bayesian method in the standard stream
Outputs:
This stream will create a directory structure that is almost the same as recon-all, but with some minor changes in the SUBJECT_DIR/mri:
native.mgz: input (reconstructed) volume.
synthseg.mgz: initial volumetric segmentation given by SynthSeg.
aseg.auto_noCCseg.mgz: same as synthseg.mgz that is required for a number of subsequent processing steps of FreeSurfer.
aseg.presurf.mgz: same as synthseg.mgz that is required for a number of subsequent processing steps of FreeSurfer.
synthSR.raw.mgz: raw output of SynthSR.
synthSR.norm.mgz: cleaned up version of synthSR.raw.mgz, scaled such that the white matter has intensity of 110.
synthsurf.mgz: file with distance maps predicted by SynthDist.
norm.mgz: generated intensity image with super-resolved cortex estimated from the distance maps in synthsurf.mgz. The subcortex of the brain is ignored here for placing surfaces.
brain.mgz: same as norm.mgz that is required for a number of subsequent processing steps of FreeSurfer.
brainmask.mgz: same as norm.mgz that is required for a number of subsequent processing steps of FreeSurfer.
wm.mgz and wm.seg.mgz: these are volumetric white matter masks used to initialize the surface placement.
Post completion of the cortical surface stream, some of the results from the cortical stream are used to refine the results in the directory SUBJECT_DIR/mri:
aseg.mgz: this is the aseg with the refined cortex label from the placed surfaces.
aparc*+aseg.mgz: same as aseg, but with parcellated cortical labels (according to 3 different atlases).
*ribbon.mgz: the ribbon, given by the surfaces.
synthSR.mgz: this is essentially synthSR.norm.mgz updated with the information from the cortical surfaces.
wmparc.mgz: white matter parcellation.
Cortical thickness and future work
The current recon-all-clinical stream is accurate for parcellation at nearly any resolution / slice spacing (see paper). However, the quality of cortical thickness estimation does degrade relatively quickly with increasing slice spacing; we plan to improve this in future versions of the tool.
