SynthSR
This functionality is only available in the development version of FreeSurfer.
Author: Juan Eugenio Iglesias
E-mail: e.iglesias [at] ucl.ac.uk
Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu
If you use this tool in your analysis, please cite:
Joint super-resolution and synthesis of 1 mm isotropic MP-RAGE volumes from clinical MRI exams with scans of different orientation, resolution and contrast. JE Iglesias, B Billot, Y Balbastre, A Tabari, J Conklin, RG Gonzalez, DC Alexander, P Golland, BL Edlow, B Fischl, for the ADNI. Neuroimage, 2021 (in press).
Since the robustness against MR contrast / orientation /resolution with a single model was not presented in the original SynthSR paper, we would also greatly appreciate it if you could please also cite:
SynthSeg: Domain Randomisation for Segmentation of Brain MRI Scans of any Contrast and Resolution. B Billot, DN Greve, O Puonti, A Thielscher, K Van Leemput, B Fischl, AV Dalca, JE Iglesias. Under revision.
Contents
- Motivation and General Description
- Installation
- Usage
- Processing Hyperfine scans
- Frequently asked questions (FAQ)
1. Motivation and General Description
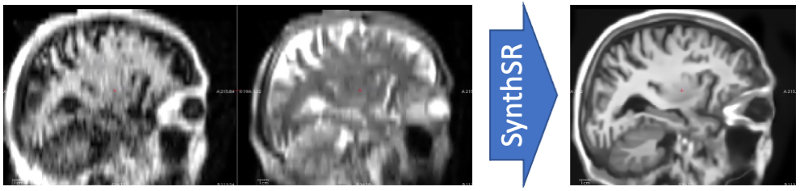
This tool implements SynthSR, a convolutional neural network that turns a clinical MRI scan (or even CT scan!) of any orientation, resolution and contrast into 1 mm isotropic MP-RAGE. You can then run your favorite neuroimaging software (including FreeSurfer, of course!) on these synthetic images for segmentation / registration / any other analysis.

2. Installation
The first time you run this module, it will prompt you to install Tensorflow. Simply follow the instructions in the screen to install the CPU or GPU version.
If you have a compatible GPU, you can install the GPU version for faster processing, but this requires installing libraries (GPU driver, Cuda, CuDNN). These libraries are generally required for a GPU, and are not specific for this tool. In fact you may have already installed them. In this case you can directly use this tool without taking any further actions, as the code will automatically run on your GPU.
3. Usage
We provide an "all purpose" model that can be applied to a scan of any resolution of contrast. Once FreeSurfer has been sourced, you can simply test SynthSR on your own data with:
synthsr --i <input> --o <output> --ct --threads <n_threads> --cpu
where:
<input>: path to an image to super-resolve / synthesize. This can also be a folder, in which case all the image inside that folder will be processed.
<output>: path where the synthetic 1 mm MP-RAGE will be saved. This must be a folder if --i designates is a folder.
--ct: (optional) use this flag when processing CT scans (details below).
<n_threads>: (optional) number of CPU threads to use. The default is just 1, so crank it up for faster processing if you have multiple cores!
--cpu: (optional) use this flag to enforce running with CPU rather than GPU.
The synthetic 1mm MP-RAGE will be of a standard contrast, bias field corrected, and with white matter lesions inpainted.
Regarding CT scans: SynthSR does a decent job with CT ! The only caveat is that the dynamic range of CT is very different to that of MRI, so they need to be clipped to [0, 80] Hounsfield units. You can use the --ct flag to do this, as long as your image volume is in Hounsfield units. If not, you will have to clip to the Hounsfield equivalent yourself (and not use --ct).
4. Processing Hyperfine scans
We also provide a dedicated, multispectral (T1+T2) model for Hyperfine scans. While you can apply the all-purpose model above to the T1 or T2 scans from Hyperfine, their relatively low in-plane resolution (1.5mm) and particularly high noise often degrades the results (especially when processing the T1). Beter results can be obtained by using both the T1 (FSE) and T2 (FSE) scans as input to a separate, dedicated version of SynthSR:
synthsr_hyperfine --t1 <t1> --t2 <t2> --o <output> --threads <n_threads> --cpu
where, as in the previous version, <t1>, <t2> and <output> can be single files or directories.
We emphasise that:
- This version is for the "standard" Hyperfine T1 and T2 acquisitions (FSE sequence) at 1.5x1.5x5mm axial resolution.
If there is motion between the T1 and T2 scans, the T2 needs to be pre-registered to the space of the T1, but without resampling to the 1.5x1.5x5mm space of the T1, which would introduce large resampling artifacts. This can be done with FreeSurfer's mri_robust_register:
mri_robust_register --mov T2.nii.gz --dst T1.nii.gz --mapmovhdr T2.reg.nii.gz --cost NMI --noinit --nomulti --lta /dev/null
- Also, we note that this Hyperfine super-resolution model is trained with synthetic images with no pathology; while it may be able to cope with small lesions, it will likely fail in e.g., stroke. We are looking forward to other super-resolution approaches (possibly supervised with real images) that do a better job!
5. Frequently asked questions (FAQ)
Does running this tool require preprocessing of the input scans?
No! Because we applied aggressive augmentation during training (see paper), this tool is able to segment both processed and unprocessed data. So there is no need to apply bias field correction, skull stripping, or intensity normalization.
What formats are supported ?
This tool can be run on Nifti (.nii/.nii.gz) and FreeSurfer (.mgz) scans.
How can I increase the speed of the CPU version without using a GPU?
If you have a multi-core machine, you can increase the number of threads with the --threads flag (up to the number of cores). Additionally you can also try to decrease the cropping value, but this will also decrease the field of view of the image.
Why aren't the predictions perfectly aligned with their corresponding images?
This is probably of problem of image viewer! Indeed, this issue might arise when performing super-resolution, when the input images and their predictions are not at the same resolution, and some viewers cannot cope with resolution changes. In that case we recommend to use the --resample flag to save the resampled images, or simply to use FreeView (shipped with FreeSurfer), which is resolution-aware !
