|
Size: 11168
Comment:
|
Size: 5605
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = SynthSeg = '''''This functionality is available in FreeSurfer development versions newer than December 2023''''' <<BR>> <<BR>> |
= WMH-SynthSeg = |
| Line 8: | Line 4: |
| ''Author: Pablo Laso'' <<BR>> |
This module is currently only available in the development version of FreeSurfer <<BR>> |
| Line 11: | Line 6: |
| ''E-mail: plaso [at] kth.se'' <<BR>> <<BR>> ''Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu'' <<BR>> <<BR>> If you use WMH-SynthSeg in your analysis, please cite: |
''Author: Pablo Laso'' <<BR>> |
| Line 19: | Line 8: |
| * [[attachment:isbi_2023_WMH-SynthSeg.pdf|Quantifying white matter hyperintensity and brain volumes in heterogeneous clinical and low-field portable MRI]]. Laso P, Cerri S, Sorby-Adams A, Guo J, Matteen F, Goebl P, Wu J, Li H, Young SI, Billot B, Puonti O, Rosen MS, Kirsch J, Strisciuglio N, Wolterink JM, Eshaghi A, Barkhof F, Kimberly WT, and Iglesias JE. Under review. | ''E-mail: plaso [at] kth.se'' <<BR>> <<BR>> ''Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu'' <<BR>> <<BR>> If you use WMH-SynthSeg in your analysis, please cite: * [[http://arxiv.org/abs/2312.05119|Quantifying white matter hyperintensity and brain volumes in heterogeneous clinical and low-field portable MRI]]. Laso P, Cerri S, Sorby-Adams A, Guo J, Matteen F, Goebl P, Wu J, Liu P, Li H, Young SI, Billot B, Puonti O, Sze G, Payabavash S, DeHavenon A, Sheth KN, Rosen MS, Kirsch J, Strisciuglio N, Wolterink JM, Eshaghi A, Barkhof F, Kimberly WT, and Iglesias JE. Proceedings of ISBI 2024 (in press). |
| Line 24: | Line 15: |
| === Contents === 1. General Description 2. Usage 3. Frequently asked questions (FAQ) |
|
| Line 25: | Line 20: |
| === Contents === | <<BR>> |
| Line 27: | Line 22: |
| 1. General Description 2. Installation 3. Usage 4. Processing CT scans 5. Frequently asked questions (FAQ) 6. Matlab implementation 7. List of segmented structures |
=== 1. General Description === This tool is a version of SynthSeg that, in addition to segmeting anatomy, also provides segmentations for white matter hyperintensity (WMH) - or hypointensities, in T1-like modalities. As the original SynthSeg, WMH-SynthSeg works out of the box and can handle brain MRI scans of any contrast and resolution. Unlike SynthSeg, WMH-SynthSeg is designed to adapt to low-field MRI scans with low resolution and signal-to-noise ratio (which makes it potentially a bit less accurate on high-resolution data acquired at high field). As for SynthSeg, the output segmentations are returned at high resolution (1mm isotropic), regardless of the resolution of the input scans. The code can run on the GPU (3s per scan) as well as the CPU (1 minute per scan). The list of segmented structures is the same as for SynthSeg 2.0 (plus the WMH label, which is FreeSurfer label 77). Below are some examples of segmentations given by WMH-SynthSeg. <<BR>> {{attachment:examples.png||height="420"}} <<BR>><<BR>> === 2. Usage === You can use WMH-SynthSeg with the following command: {{{ mri_WMHsynthseg --i <input> --o <output> [--csv_vols <CSV file>] [--device <device>] [--threads <threads>] [--crop] [--save_lesion_probabilities] }}} where: * ''<input>'': path to a scan to segment, or to a folder. * ''<output>'': path where the output segmentations will be saved. This must be the same type as ''--i'' (i.e., the path to a file or a folder). * ''<CSV file>'': (optional) path to a CSV file where volumes for all segmented regions will be saved. * ''<threads>'': (optional) number of threads to be used by PyTorch in CPU mode (see blow). Default is 1. Set it to -1 to use all possible cores. * ''<device>'': (optional) device used by PyTorch. Default is 'cpu'. Set it to 'cuda' to use the GPU, if available. If there are multiple GPUs available, use 'cuda:0', 'cuda:1', etc to index them. * ''<crop>'': (optional) runs 2 passes of the algorithm: one to roughly find the center of the brain, and another to do the segmentation processing only a portion of the image, cropped around the center. You will need to activate this option if you are using a GPU (long story). * ''<save_lesion_probabilities>'': (optional) saves additional files with the soft segmentations of the WMHs. '''''Important:''''' If you wish to process several scans, we highly recommend that you put them in a single folder, rather than calling mri_WMHsynthseg individually on each scan. This will save the time required to set up the software for each scan. |
| Line 38: | Line 51: |
| === 1. General Description === | === 3. Frequently asked questions (FAQ) === * '''What are the computation requirements for this tool?''' |
| Line 40: | Line 54: |
| This tool is a version of SynthSeg that, in addition to segmeting anatomy, also provides segmentations for white matter hyperintensity (WMH) - or hypointensities, in T1-like modalities. As the original SynthSeg, WMH-SynthSeg works out of the box and can handle brain MRI scans of any contrast and resolution. Unlike SynthSeg, WMH-SynthSeg is designed to adapt to low-field MRI scans with low resolution and signal-to-noise ratio (which makes it potentially a bit less accurate on high-resolution data acquired at high field). As for SynthSeg, the output segmentations are returned at high resolution (1mm isotropic), regardless of the resolution of the input scans. The code can run on the GPU (3s per scan) as well as the CPU (1 minute per scan). The list of segmented structures is the same as for SynthSeg (plus the WMH label, which is FreeSurfer label 77). <<BR>> {{attachment:examples.png||height="320"}} <<BR>><<BR>> <<BR>> === New features (28/06/2022) === SynthSeg 1.0 has now been replaced with SynthSeg 2.0, which offers more functionalities !! These new features are illustrated in the figure below. Specifically, in addition to whole-brain segmentation, SynthSeg is now also able to perform cortical parcellation, automated quality control (QC) of the produced segmentations, and intracranial volume (ICV) estimation computed by including the CSF to the list of segmented structures. <<BR>> {{attachment:new_features2.png||height="320"}} <<BR>> === Robust version === While SynthSeg is quite robust, it sometimes falters on scans with low signal-to-noise ratio, or with low tissue contrast (see figure below). For this reason, we developed a new architecture for increased robustness, named "SynthSeg-robust", which can be selected with the --robust flag (see Section 3). You can use this mode when SynthSeg gives results like those in the third column (1st and 2nd rows) of the figure below: <<BR>> {{attachment:robust2.png||height="320"}} <<BR>> See the table below for a summary of the functionalities supported by each version. <<BR>> {{attachment:table_versions2.png||height="180"}} <<BR>> === 2. Installation === The first time you run this module, it will prompt you to install Tensorflow. Simply follow the instructions in the screen to install the CPU or GPU version. If you have a compatible GPU, you can install the GPU version for faster processing, but this requires installing libraries (GPU driver, Cuda, CuDNN). These libraries are generally required for a GPU, and are not specific for this tool. In fact you may have already installed them. In this case you can directly use this tool without taking any further actions, as the code will automatically run on your GPU. <<BR>> === 3. Usage === You can use SynthSeg with the following command: {{{ mri_synthseg --i <input> --o <output> [--parc --robust --fast --vol <vol> --qc <qc> --post <post> --resample <resample> --crop <crop> --threads <threads> --cpu --v1 --ct] }}} where: * ''<input>'': path to a scan to segment, or to a folder. This can also be the path to a text file, where each line is the path of an image to segment. * ''<output>'': path where the output segmentations will be saved. This must be the same type as ''--i'' (i.e., the path to a file, a folder, or a text file where each line is the path to an output segmentation). * ''--parc'': (optional) to perform cortical parcellation in addition to whole-brain segmentation. * ''--robust'': (optional) to use the variant for increased robustness (e.g., when analysing clinical data with large space spacing). This can be slower than the other model. * ''--fast'': (optional) use this flag to disable some postprocessing operations for faster prediction (approximately twice as fast, but slightly less accurate). This doesn't apply when the --robust flag is used. * ''<vol>'': (optional) path to a CSV file where volumes for all segmented regions will be saved. If ''--i'' designates an image or a folder, ''<vol>'' must be the path to a single CSV file where the volumes of all subjects will be saved. Otherwise, ''<vol>'' must be a text file, where each line is the path to a different CSV file where the volumes of the corresponding subject will be saved. * ''<qc>'': (optional) path to a CSV file where QC scores will be saved. The same formatting requirements apply as for the --vol flag. * ''<post>'': (optional) path where the output 3D posterior probabilities will be saved. This must be the same type as ''--i'' (i.e., the path to a file, a folder, or a text file). * ''<resample>'': (optional) in order to return segmentations at 1mm resolution, the input images are internally resampled (except if they already are at 1mm). Use this optional flag to save the resampled images. This must be the same type as ''--i'' (i.e., the path to a file, a folder, or a text file). * ''<crop>'': (optional) to crop the inputs to a given shape before segmentation. This must be divisible by 32. It can be given as a single (i.e., `--crop 160`) or several integers (i.e, `--crop 160 128 192`, ordered in RAS coordinates). By default the whole image is processed. Use this flag for faster analysis or to fit in your GPU. * ''<threads>'': (optional) number of threads to be used by Tensorflow (default uses one core). Increase it to decrease the runtime when using the CPU version. * ''--cpu'': (optional) to run on the CPU rather than the GPU. * ''--v1'': (optional) to run the first version of SynthSeg (SynthSeg 1.0). * ''--ct'': (optional) use this flag when processing CT scans (details below). * ''--photo'': (optional) use this flag when processing stacks of 3D reconstructed dissection photos (see PhotoTools). Use --photo left to segment left hemispheres, --photo right for right hemispheres, and --photo both for images with both hemispheres. We note that '''--parc''', '''--robust''', '''--fast''', '''--qc''', '''--v1''', as well as '''input text files''', are only available on development versions from '''June 28th 2022 onwards'''. '''''Important:''''' If you wish to process several scans, we highly recommend that you put them in a single folder, or use input text files, rather than calling SynthSeg individually on each scan. This will save the time required to set up the software for each scan. <<BR>> === 4. Processing CT scans === Regarding CT scans: SynthSR does a decent job with CT ! The only caveat is that the dynamic range of CT is very different to that of MRI, so they need to be clipped to [0, 80] Hounsfield units. You can use the --ct flag to do this, as long as your image volume is in Hounsfield units. If not, you will have to clip to the Hounsfield equivalent yourself (and not use --ct). === 5. Frequently asked questions (FAQ) === |
About 32GB of RAM memory. |
| Line 130: | Line 58: |
| No! Because we applied aggressive augmentation during training (see paper), this tool is able to segment both processed and unprocessed data. So there is no need to apply bias field correction, skull stripping, or intensity normalisation. |
No! Because we applied aggressive augmentation during training (see paper), this tool is able to segment both processed and unprocessed data. So there is no need to apply bias field correction, skull stripping, or intensity normalization. |
| Line 136: | Line 62: |
| This is because the volumes are computed upon a soft segmentation, rather than the discrete segmentation. The same happens with the main recon-all stream: if you compute volumes by counting voxels in aseg.mgz, you don't get the values reported in aseg.stats. |
This is because the volumes are computed upon a soft segmentation, rather than the discrete segmentation. The same happens with the main recon-all stream: if you compute volumes by counting voxels in aseg.mgz, you don't get the values reported in aseg.stats. |
| Line 144: | Line 68: |
| Line 147: | Line 70: |
| The first solution is to use the --fast flag, which will half the processing time if you're not using the "--robust" flag (gains in speed are much smaller if you are). Next, if you have a multi-core machine, you can increase the number of threads with the --threads flag (up to the number of cores). Additionally you can also try to decrease the cropping value, but this will also decrease the field of view of the image. |
If you have a multi-core machine, you can increase the number of threads with the --threads flag (up to the number of cores). |
| Line 154: | Line 74: |
| Simply because, in order to output segmentations at 1mm resolution, the network needs the input images to be at this particular resolution! We actually do not resample images with resolution in the range [0.95, 1.05], which is close enough. We highlight that the resampling is performed internally to avoid the dependence on any external tool. |
Simply because, in order to output segmentations at 1mm resolution, the network needs the input images to be at this particular resolution! We highlight that the resampling is performed internally to avoid the dependence on any external tool. |
| Line 161: | Line 78: |
| This may happens with viewers other than FreeSurfer's Freeview, if they do not handle headers properly. We recommend using Freeview but, if you want to use another viewer, you may have to use the ''--resample'' flag to save the resampled images, which any viewer will correctly align with the segmentations. |
This may happens with viewers other than FreeSurfer's Freeview, if they do not handle headers properly. We recommend using Freeview but, if you want to use another viewer, you may can use mri_convert with the -rl flag to obtain resampled images, which any other view will display correctly. Something like: 'mri_convert input.nii.gz input.resampled.nii.gz -rl segmentation.nii.gz'. |
| Line 165: | Line 81: |
=== 6. Matlab implementation === Matlab added the non-robust version of SynthSeg to their Medical Imaging Toolbox in version R2022b. They have a fully documented example on how to use it [[https://www.mathworks.com/help/medical-imaging/ug/Brain-MRI-Segmentation-Using-Trained-3-D-U-Net.html|here]]. Alternatively, you can download our Matlab script, which you can call with a single line of code: [[https://surfer.nmr.mgh.harvard.edu/fswiki/SynthSeg?action=AttachFile&do=get&target=SynthSegMatlab.zip|Download]]. Uncompress the code and type: "help SynthSeg" for instructions. === 7. List of segmented structures === Please note that the label values follow the FreeSurfer classification. We emphasise that the structures are given in the same order as they appear in the posteriors, i.e. the first map of the posteriors corresponds to the background, then the second map is associated to the left cerebral white matter, etc. <<BR>> {{attachment:table_labels2.png||height="550"}} |
WMH-SynthSeg
This module is currently only available in the development version of FreeSurfer
Author: Pablo Laso
E-mail: plaso [at] kth.se
Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu
If you use WMH-SynthSeg in your analysis, please cite:
Quantifying white matter hyperintensity and brain volumes in heterogeneous clinical and low-field portable MRI. Laso P, Cerri S, Sorby-Adams A, Guo J, Matteen F, Goebl P, Wu J, Liu P, Li H, Young SI, Billot B, Puonti O, Sze G, Payabavash S, DeHavenon A, Sheth KN, Rosen MS, Kirsch J, Strisciuglio N, Wolterink JM, Eshaghi A, Barkhof F, Kimberly WT, and Iglesias JE. Proceedings of ISBI 2024 (in press).
Contents
- General Description
- Usage
- Frequently asked questions (FAQ)
1. General Description
This tool is a version of SynthSeg that, in addition to segmeting anatomy, also provides segmentations for white matter hyperintensity (WMH) - or hypointensities, in T1-like modalities. As the original SynthSeg, WMH-SynthSeg works out of the box and can handle brain MRI scans of any contrast and resolution. Unlike SynthSeg, WMH-SynthSeg is designed to adapt to low-field MRI scans with low resolution and signal-to-noise ratio (which makes it potentially a bit less accurate on high-resolution data acquired at high field).
As for SynthSeg, the output segmentations are returned at high resolution (1mm isotropic), regardless of the resolution of the input scans. The code can run on the GPU (3s per scan) as well as the CPU (1 minute per scan). The list of segmented structures is the same as for SynthSeg 2.0 (plus the WMH label, which is FreeSurfer label 77). Below are some examples of segmentations given by WMH-SynthSeg.
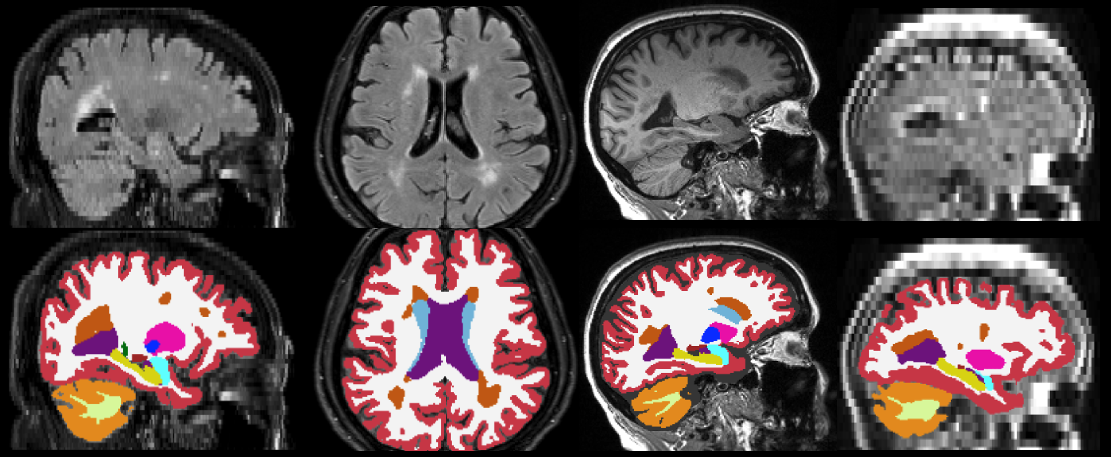
2. Usage
You can use WMH-SynthSeg with the following command:
mri_WMHsynthseg --i <input> --o <output> [--csv_vols <CSV file>] [--device <device>] [--threads <threads>] [--crop] [--save_lesion_probabilities]
where:
<input>: path to a scan to segment, or to a folder.
<output>: path where the output segmentations will be saved. This must be the same type as --i (i.e., the path to a file or a folder).
<CSV file>: (optional) path to a CSV file where volumes for all segmented regions will be saved.
<threads>: (optional) number of threads to be used by PyTorch in CPU mode (see blow). Default is 1. Set it to -1 to use all possible cores.
<device>: (optional) device used by PyTorch. Default is 'cpu'. Set it to 'cuda' to use the GPU, if available. If there are multiple GPUs available, use 'cuda:0', 'cuda:1', etc to index them.
<crop>: (optional) runs 2 passes of the algorithm: one to roughly find the center of the brain, and another to do the segmentation processing only a portion of the image, cropped around the center. You will need to activate this option if you are using a GPU (long story).
<save_lesion_probabilities>: (optional) saves additional files with the soft segmentations of the WMHs.
Important: If you wish to process several scans, we highly recommend that you put them in a single folder, rather than calling mri_WMHsynthseg individually on each scan. This will save the time required to set up the software for each scan.
3. Frequently asked questions (FAQ)
What are the computation requirements for this tool?
About 32GB of RAM memory.
Does running this tool require preprocessing of the input scans?
No! Because we applied aggressive augmentation during training (see paper), this tool is able to segment both processed and unprocessed data. So there is no need to apply bias field correction, skull stripping, or intensity normalization.
The sum of the number of voxels of a given structure multiplied by the volume of a voxel is not equal to the volume reported in the output volume file.
This is because the volumes are computed upon a soft segmentation, rather than the discrete segmentation. The same happens with the main recon-all stream: if you compute volumes by counting voxels in aseg.mgz, you don't get the values reported in aseg.stats.
What formats are supported ?
This tool can be run on Nifti (.nii/.nii.gz) and FreeSurfer (.mgz) scans.
How can I increase the speed of the CPU version without using a GPU?
If you have a multi-core machine, you can increase the number of threads with the --threads flag (up to the number of cores).
Why are the inputs automatically resampled to 1mm resolution ?
Simply because, in order to output segmentations at 1mm resolution, the network needs the input images to be at this particular resolution! We highlight that the resampling is performed internally to avoid the dependence on any external tool.
Why aren't the segmentations perfectly aligned with their corresponding images?
This may happens with viewers other than FreeSurfer's Freeview, if they do not handle headers properly. We recommend using Freeview but, if you want to use another viewer, you may can use mri_convert with the -rl flag to obtain resampled images, which any other view will display correctly. Something like: 'mri_convert input.nii.gz input.resampled.nii.gz -rl segmentation.nii.gz'.
