|
Size: 35
Comment:
|
Size: 3643
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| Describe mri_3d_photo_recon here. | = Co-registration of dissection photographs of coronal slabs into a 3D volume = '''''For FreeSurfer 8.0 or development versions downloaded before May 2025, please see [[https://surfer.nmr.mgh.harvard.edu/fswiki/mri_3d_photo_recon_2024]].''''' If you use this tools in your analysis, please cite: * [[https://elifesciences.org/articles/91398|Machine learning of dissection photographs and surface scanning for quantitative 3D neuropathology]]. H Gazula, H Tregidgo, B Billot, Y Balbastre, J Williams Ramirez, R Herisse, LJ Deden-Binder, A Casamitjana, EJ Melief, CS Latimer, MD Kilgore, M Montine, E Robinson, E Blackburn, MS Marshall, TR Connors, DH Oakley, MP Frosh, SI Young, K Van Leemput, AV Dalca, B Fischl, CL Mac Donald, CD Keene, B Hyman, and JE Iglesias. Under revision.) This is a newer version of our [[https://surfer.nmr.mgh.harvard.edu/fswiki/mri_3d_photo_recon_2024|older tool]] with several key improvements: * More flexible framework, accepting a variable number of references (e.g., surface scan and/or MRI scan or MNI atlas etc). * Faster optimization * Easier configuration of parameters: it self configures depending on type(s) of input(s) * Supports slabs of variable thickness * New feature: machine learning interpolation to 1mm slab thickness (fun stuff! See pictures below) mri_3d_photo_recon -h ||--input_photo_dir ||Directory with input pixel-corrected photos. || ||--input_segmentation_dir ||Directory with input slab masks/segmentations || ||--ref_mask ||When using a binary volume as a reference. || ||--ref_surface ||Using a 3D surface scan as a reference. || ||--ref_soft_mask ||Using the provided average atlas. Best for using retrospective processing on data without a better reference. || ||--mesh_reorient_with_indices ||Vertex indices of the frontal pole, occipital pole, and top of the central sulcus, separated with commas, for mesh alignment. || ||--photos_of_posterior_side ||Use when photos are taken of the posterior side of slabs (default is anterior side). || ||--order_posterior_to_anterior ||Use when photos are ordered from posterior to anterior (default is anterior to posterior). || ||--allow_z_stretch ||Use to adjust the slice thickness to best match the reference. You should probably '''*never*''' use this with soft references (ref_soft_mask). || ||--rigid_only_for_photos ||Switch on if you want photos to deform only rigidly (not affine). || ||--slice_thickness ||Slice thickness in mm. || ||--photo_resolution ||Resolution of the photos in mm. || ||--output_directory ||Output directory with reconstructed photo volume and reference || <<BR>> In Freeview, you can find the number corresponding to the vertices of each anatomical area to use in the --mesh_reorient_with_indices flag. These indices should be comma-separated in the order: frontal pole, occipital pole, and top of the central sulcus.<<BR>> || {{attachment:43_vertexFP.png||width="304",height="254"}} || {{attachment:44_vertexOP.png||width="297",height="249"}} || {{attachment:45_vertexCS.png||width="302",height="253"}} || ||Frontal Pole ||Occipital Pole ||Top of the central sulcus || For example, the --mesh_reorient_with_indices flag might look like this: . –mesh_reorient_with_indices 999999,999999,999999 '''Example for running reconstruction''' <<BR>> . mri_3d_photo_recon \ . --input_photo_dir ./deformed \ . --input_segmentation_dir ./connected_components \ . --ref_surface ./mesh/case.stl \ . --mesh_reorient_with_indices 999,1010,333333 \ . --photos_of_posterior_side --allow_z_stretch \ . --slice_thickness 8 \ . --photo_resolution 0.1 |
Co-registration of dissection photographs of coronal slabs into a 3D volume
For FreeSurfer 8.0 or development versions downloaded before May 2025, please see https://surfer.nmr.mgh.harvard.edu/fswiki/mri_3d_photo_recon_2024.
If you use this tools in your analysis, please cite:
* Machine learning of dissection photographs and surface scanning for quantitative 3D neuropathology. H Gazula, H Tregidgo, B Billot, Y Balbastre, J Williams Ramirez, R Herisse, LJ Deden-Binder, A Casamitjana, EJ Melief, CS Latimer, MD Kilgore, M Montine, E Robinson, E Blackburn, MS Marshall, TR Connors, DH Oakley, MP Frosh, SI Young, K Van Leemput, AV Dalca, B Fischl, CL Mac Donald, CD Keene, B Hyman, and JE Iglesias. Under revision.)
This is a newer version of our older tool with several key improvements:
- More flexible framework, accepting a variable number of references (e.g., surface scan and/or MRI scan or MNI atlas etc).
- Faster optimization
- Easier configuration of parameters: it self configures depending on type(s) of input(s)
- Supports slabs of variable thickness
- New feature: machine learning interpolation to 1mm slab thickness (fun stuff! See pictures below)
mri_3d_photo_recon -h
--input_photo_dir |
Directory with input pixel-corrected photos. |
--input_segmentation_dir |
Directory with input slab masks/segmentations |
--ref_mask |
When using a binary volume as a reference. |
--ref_surface |
Using a 3D surface scan as a reference. |
--ref_soft_mask |
Using the provided average atlas. Best for using retrospective processing on data without a better reference. |
--mesh_reorient_with_indices |
Vertex indices of the frontal pole, occipital pole, and top of the central sulcus, separated with commas, for mesh alignment. |
--photos_of_posterior_side |
Use when photos are taken of the posterior side of slabs (default is anterior side). |
--order_posterior_to_anterior |
Use when photos are ordered from posterior to anterior (default is anterior to posterior). |
--allow_z_stretch |
Use to adjust the slice thickness to best match the reference. You should probably *never* use this with soft references (ref_soft_mask). |
--rigid_only_for_photos |
Switch on if you want photos to deform only rigidly (not affine). |
--slice_thickness |
Slice thickness in mm. |
--photo_resolution |
Resolution of the photos in mm. |
||--output_directory ||Output directory with reconstructed photo volume and reference ||
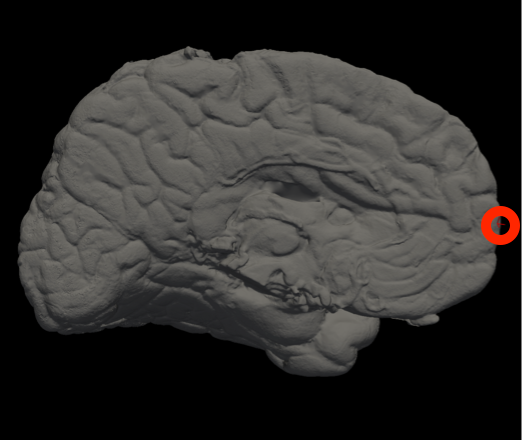
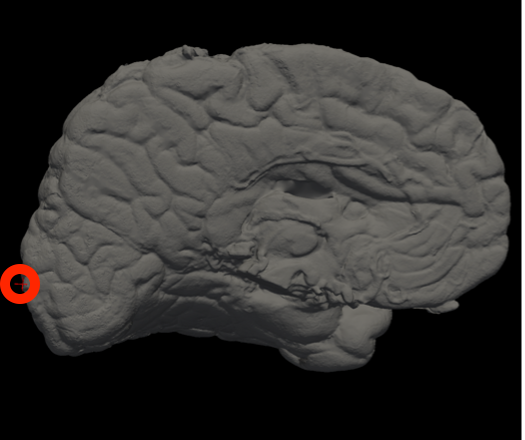
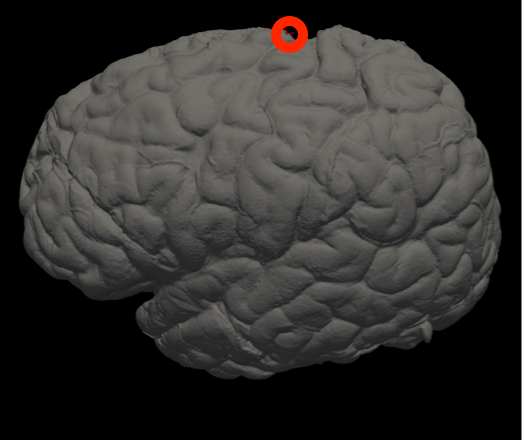
In Freeview, you can find the number corresponding to the vertices of each anatomical area to use in the --mesh_reorient_with_indices flag. These indices should be comma-separated in the order: frontal pole, occipital pole, and top of the central sulcus.
|
|
|
Frontal Pole |
Occipital Pole |
Top of the central sulcus |
For example, the --mesh_reorient_with_indices flag might look like this:
- –mesh_reorient_with_indices 999999,999999,999999
Example for running reconstruction
- mri_3d_photo_recon \
- --input_photo_dir ./deformed \
- --input_segmentation_dir ./connected_components \
- --ref_surface ./mesh/case.stl \
- --mesh_reorient_with_indices 999,1010,333333 \
- --photos_of_posterior_side --allow_z_stretch \
- --slice_thickness 8 \
- --photo_resolution 0.1