| Deletions are marked like this. | Additions are marked like this. |
| Line 25: | Line 25: |
| {{attachment:sample_output_recon_all_clinical.png||width="720",height="405"}} <<BR>>''Out of the box cortical surface reconstruction and analysis of heterogenous scans. (a)Sagittal T1 scan with .4×.4×6mm resolution. (b)Axial FLAIR scan with 1.7×1.7×6mm resolution. (c)Axial T2-weighted scan with .9×.9×6mm resolution .''<<BR>> | {{attachment:sample_output_recon_all_clinical.png||height="405",width="720"}} <<BR>>''Out of the box cortical surface reconstruction and analysis of heterogenous scans. (a)Sagittal T1 scan with .4×.4×6mm resolution. (b)Axial FLAIR scan with 1.7×1.7×6mm resolution. (c)Axial T2-weighted scan with .9×.9×6mm resolution. The WM surface with cortical parcellation overlaid and pial surfaces are also shown.''<<BR>> |
recon-all-clinical
This functionality is now available in the developer version of FreeSurfer.
Author: Karthik Gopinath
E-mail: kgopinath[at]mgh[dot]harvard[dot]edu
Please post your questions on this module to the FreeSurfer mailing list at freesurfer[at]nmr.mgh.harvard.edu rather than directly contacting the author.
If you use this package in your analysis, please cite:
Cortical analysis of heterogeneous clinical brain MRI scans for large-scale neuroimaging studies. K Gopiath, DN Greeve, S Das, S Arnold, C Magdamo, JE Iglesias
Joint super-resolution and synthesis of 1 mm isotropic MP-RAGE volumes from clinical MRI exams with scans of different orientation, resolution and contrast. JE Iglesias, B Billot, Y Balbastre, A Tabari, J Conklin, RG Gonzalez, DC Alexander, P Golland, BL Edlow, B Fischl, for the ADNI. Neuroimage, 118206 (2021).
SynthSR: a public AI tool to turn heterogeneous clinical brain scans into high-resolution T1-weighted images for 3D morphometry. JE Iglesias, B Billot, Y Balbastre, C Magdamo, S Arnold, S Das, B Edlow, D Alexander, P Golland, B Fischl. Science Advances, 9(5), eadd3607 (2023).
General description:
This tool performs recon-all-clinical, the first out-of-the-box cortical surface reconstruction and analysis of brain MRI scans of any modality, contrast and resolution without retraining and fine-tuning.
This "Recon-all-like" stream for clinical scans of arbitrary orientation/resolution/contrast is essentially a combination of:
SynthSeg: to obtain an aseg.auto_noCCseg.mgz and to compute a Talairach transform
SynthSR: to have a higher resolution 1mm MPRAGE for visualization
SynthSurfaces: to fit surfaces by predicting the distance maps and reconstructing topologically accurate cortical surfaces
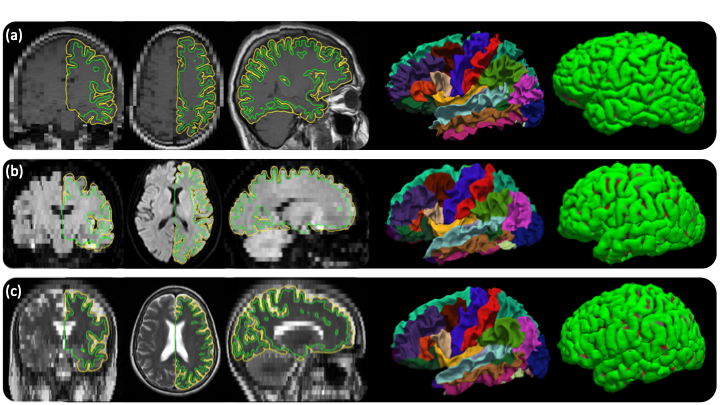
Out of the box cortical surface reconstruction and analysis of heterogenous scans. (a)Sagittal T1 scan with .4×.4×6mm resolution. (b)Axial FLAIR scan with 1.7×1.7×6mm resolution. (c)Axial T2-weighted scan with .9×.9×6mm resolution. The WM surface with cortical parcellation overlaid and pial surfaces are also shown.
Usage:
OnceFreeSurfer has been sourced, you can simply run recon-all-clinical on your own data with
recon-all-clinical.sh INPUT_SCAN SUBJECT_ID THREADS [SUBJECT_DIR]
where:
- INPUT_SCAN: path to an image that will be processed.
- SUBJECT_ID: specifies the name or ID of the subject you would like to use. A directory with that name will be created for all the subject's FreeSurfer output.
- THREADS (optional): number of CPU threads to use. The default is just 1, so crank it up for faster processing if you have multiple cores!
- SUBJECT_DIR: only necessary if the environment variable SUBJECTS_DIR has not been set when sourcing FreeSurfer or if you want to override it.
This stream runs a bit faster than the original recon-all, since the volumetric segmentation is much faster than the iterative Bayesian method in the standard stream
