TRACULA (TRActs Constrained by UnderLying Anatomy) is a tool for automatic reconstruction of a set of major white-matter pathways from diffusion-weighted MRI data. As the FreeSurfer cortical parcellation and subcortical segmentation use prior knowledge on the relative positions of anatomical structures with respect to each other, TRACULA uses prior knowledge on the relative positions of white-matter pathways with respect to their surrounding anatomical structures. Whereas the anatomical segmentation/parcellation tools use this type of neighborhood information to classify voxels in the volume or vertices on the surface, TRACULA uses it to produce tractography streamlines.
Features
TRACULA uses global probabilistic tractography with anatomical neighborhood priors. Prior distributions on the neighboring anatomical structures of each white-matter pathway are derived from a set of manually annotated training subjects. When TRACULA reconstructs a pathway of interest in a novel subject, it fits the shape of the pathway both to the fiber orientation vectors derived from the subject's diffusion MRI data, and to the anatomical neighborhood priors derived from the subject's T1 data. This obviates the need for any user interaction, such as having to define ad hoc regions of interest or thresholds on curvature and length for each pathway.
In addition to this core functionality, TRACULA provides an end-to-end pipeline that allows the user to:
- Preprocess the diffusion-weighted images, applying standard methods to mitigate image distortions due to eddy currents and B0 field inhomogeneities
- Extract measures of head motion that can be used as co-variates in subsequent statistical analyses
- Register the individual's diffusion-weighted and anatomical images to each other
- Register the individual to a common (template) space
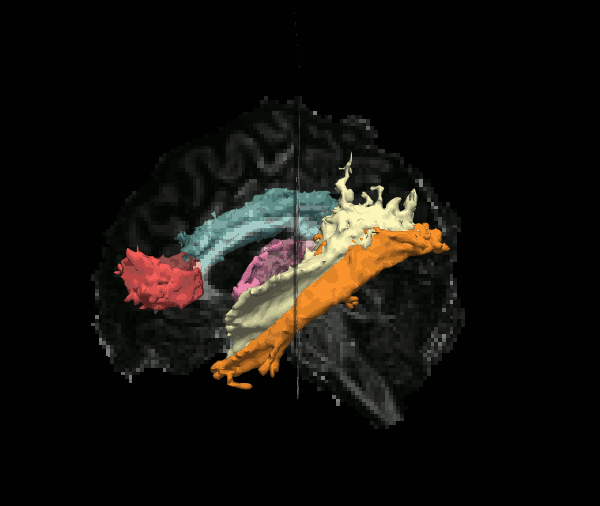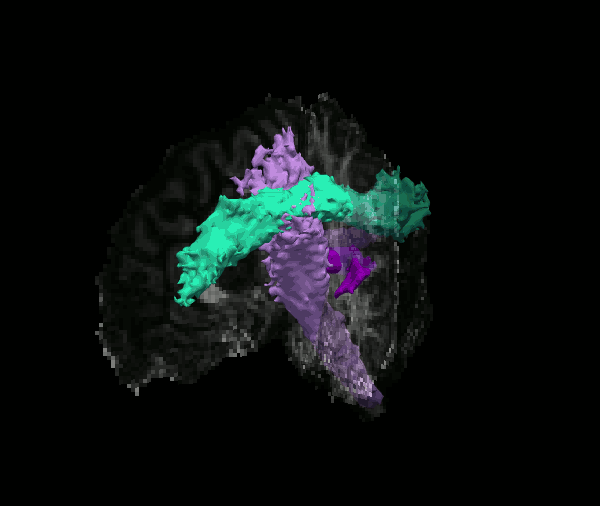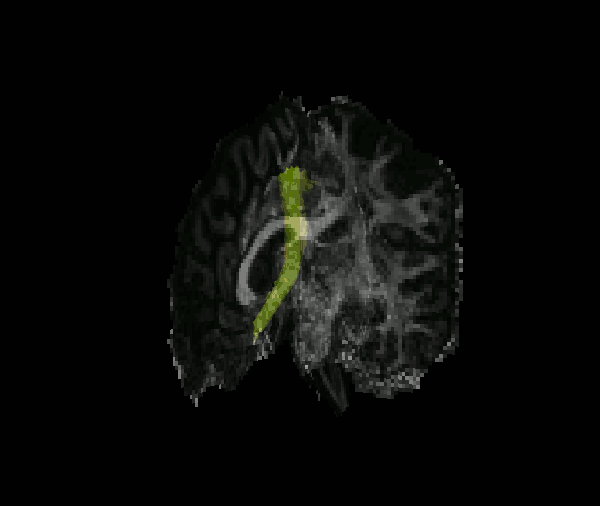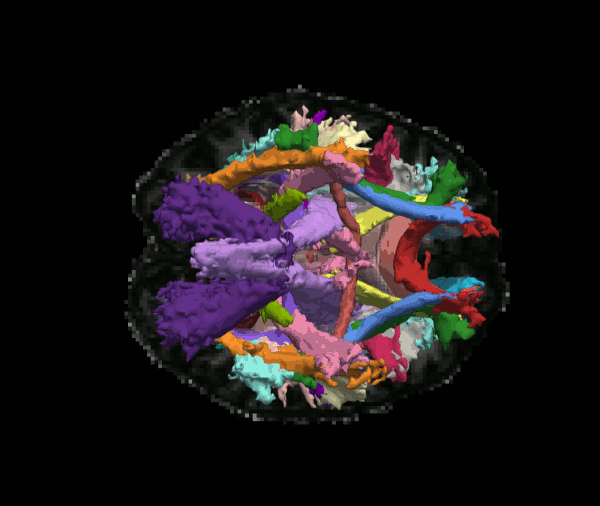
Reconstruct probability distributions of 42 major white-matter pathways
Extract standard diffusion measures (FA, MD, etc.) for each of the reconstructed pathways, either as average values over each pathway or as profiles along the trajectory of each pathway
- Perform statistics on the above measures and visualize the results
Finally, TRACULA offers a dedicated stream for processing longitudinal diffusion MRI data. The longitudinal stream reconstructs the pathways of interest jointly from a subject's data at all time points, rather than processing each time point independently as if it were a cross-sectional data point.
Getting started
The best way to get started using TRACULA is by following the tutorials from the FreeSurfer training workshop:
A somewhat older version of the slide presentations from the workshop is available here:
Note that, because TRACULA relies on the underlying anatomy as derived from the FreeSurfer cortical parcellation and subcortical segmentation, these need to be generated first. This means that, before analyzing your diffusion MRI data with TRACULA, you will have to analyze your subjects' T1 data with recon-all and make sure that they have a good-quality mri/aparc+aseg.mgz. In addition to this whole-brain segmentation, the latest version of TRACULA can make use of the thalamic nuclei segmentation, to improve reconstruction of pathways that terminate in or neighbor the thalamus. Thus it is recommended that, prior to TRACULA, you also run the thalamic nuclei segmentation, which will save its output as mri/ThalamicNuclei.v12.T1.FSvoxelSpace.mgz.
Documentation
All preprocessing, tractography, and post-processing steps are performed by the trac-all script. Several options for each analysis step can be set by the user in a configuration file (dmrirc file), which is passed as an argument to trac-all.
More detailed documentation is available for both trac-all and the configuration files, in case you need more customization of processing options or more information on all the outputs:
trac-all usage (including description of all processing steps)
White-matter tract atlas
The TRACULA atlas is included in the FreeSurfer distribution and currently comprises 42 major pathways.
Although we refer to it as an "atlas", it is not simply an average brain where voxels have been labeled as tracts. It consists of both tractography streamlines and anatomical segmentations from individual subjects. These subjects were imaged on the MGH Connectom scanner, as part of the young adult Human Connectome Project.
Updates
TRACULA in FreeSurfer 7.3 (upcoming):
- Auto-configure TRACULA to run on BIDS-formatted datasets
TRACULA in FreeSurfer 7.2:
New set of 42 white-matter pathways. TRACULA is now trained on manual annotations of these 42 pathways from the MGH Connectom data, and can reconstruct these pathways automatically in much lower quality (e.g., b=1000, 32- or 64-direction) data. For more info, see our paper.
Added support for multiple diffusion scans per scan session (with same or different direction sets, with or without reverse polarity). See new example configuration files ($FREESURFER_HOME/bin/dmrirc.multiscan.example for cross-sectional, $FREESURFER_HOME/bin/dmrirc.long.multiscan.example for longitudinal).
Added pre-processing options to use eddy for eddy-current distortion correction (now the default) and/or topup for susceptibility distortion correction.
Added more options for registration to a common coordinate system (robust affine with mri_robust_register or nonlinear with ANTs). Nonlinear registration uses a custom FA template, built from the MGH Connectom data.
Refactored the anatomical prior computation (-prior step), which is now an order of magnitude faster.
Added bshell and bmax configuration options that allow processing steps to be run on a specific subset of the data (e.g., to compute tensor measures from a single shell out of a multi-shell scan).
Pointwise Assessment of Streamline Tractography Attributes (PASTA), a.k.a., along-tract measures can be fed directly to mri_glmfit(-sim) for statistical analysis with FSGD files, just like surface- or volume-based maps.
- The statistical maps from PASTA analysis can be projected onto .trk files for visualization in freeview.
The cortical end points of each reconstructed pathway are projected onto surface labels, and anatomical stats (e.g., cortical thickness) are extracted for further analysis.
TRACULA in FreeSurfer 6.0:
- Automatic path reinitialization: This feature reduces the cases where the user has to reinitialize a path dramatically.
Cluster job submission: Adding -jobs <filename> to the trac-all command line will process the configuration file, set up the analysis for all subjects included in it, but will not actually run the analysis. Instead, it will write the commands that need to be run into <filename>. The user is then responsible for submitting those as jobs to be run in parallel on a cluster.
Gradient tables and b-value tables: They can now be read directly from the DICOM header of the DWI series (using new functionality in mri_convert), without the user having to specify them.
In case you want to remove certain volumes from your DWIs, you can create a text file called dmri/keepframes.txt for each subject, containing the list of frames (0-based) that you want to keep. In this case, you provide the entire DWI series (and the entire gradient table and b-value table, if they are not to be read from the DICOM header) but TRACULA only uses the subset of frames specified in this file.
It is also possible to specify a different table of b-values for each scan, using set bvallist instead of set bvalfile in the configuration file.
TRACULA in FreeSurfer 5.3:
2014/05/26 --
 TRACULA update: CentOS 6 (64-bit) | CentOS 4 (64-bit) | CentOS 4 (32-bit) | MacOSX lion (64-bit) | MacOSX snow leopard (32-bit) This update includes all the features from the previous update, plus improvements to the longitudinal stream of TRACULA. Note that this newer version of longitudinal TRACULA uses the aparc+aseg.mgz from the *.long.* output of longitudinal FreeSurfer, rather than the cross-sectional aparc+aseg.mgz.
TRACULA update: CentOS 6 (64-bit) | CentOS 4 (64-bit) | CentOS 4 (32-bit) | MacOSX lion (64-bit) | MacOSX snow leopard (32-bit) This update includes all the features from the previous update, plus improvements to the longitudinal stream of TRACULA. Note that this newer version of longitudinal TRACULA uses the aparc+aseg.mgz from the *.long.* output of longitudinal FreeSurfer, rather than the cross-sectional aparc+aseg.mgz. - 2013/12/09
NEW FEATURE: Compute the four measures of head motion from Yendiki et al. 2013, which can be used to match motion between groups or regress motion in statistical analyses. This can be run as a separate step with trac-all -qa, and is also run as part of trac-all -prep.
NEW FEATURE: Combine anisotropy and diffusivity measures from multiple subjects at different positions along each pathway (from the pathstats.byvoxel.txt files). This can be run with trac-all -stat.
It is now possible to specify a different table of gradient vectors for each scan, using set bveclist instead of set bvecfile in the configuration file.
The tables of gradient vectors and/or b-values can now be formatted in column or row format, and blank lines at the end of each file should not cause an error.
- A scripting bug that was causing the longitudinal stream not to run at all on certain platforms has been fixed.
2013/05/15 -- No changes to TRACULA in FreeSurfer 5.3 since FreeSurfer 5.2
TRACULA in FreeSurfer 5.2:
2013/03/02 -- Changes to TRACULA included in FreeSurfer 5.2:
- Improved fitting for the initialization of the pathways in the -prior step, along with a 70% reduction in computation time for that step.
- More efficient exploration of 3D space in the pathway reconstruction algorithm allows it to get to more of the pathway endings and those hard-to-reach spots.
- Several default settings have changed, see $FREESURFER_HOME/bin/dmrirc.example for details:
- The default and recommended method for intra-subject registration is now bbregister.
- A different number of control points can be (and is, by default) chosen for each of the 18 pathways.
- The default number of samples has increased.
The CVS template can now be used instead of the MNI template for inter-subject registration. This could improve the initialization of the pathways for subjects with challenging anatomical features, e.g., enlarged ventricles, at the expense of higher memory requirements for holding the CVS non-linear warp in memory.
- The low-b images can now be interspersed throughout the DWI series and the low b-value can be greater than zero. There is no need for the user to define the number of low-b images in the configuration file any more, this will all be worked out automatically from the b-value table.
- The user can set the number of anisotropic compartments ("sticks") in bedpostx's ball-and-stick model to something other than 2, by setting the nstick variable in the configuration file.
- Compatibility issues with the command line options of the latest version of bedpostx have been resolved.
The maximum a posteriori path, which is used to produce statistics along the length of the path (pathstats.byvoxel.txt) is now selected in a more robust way that makes these paths more consistent across subjects.
In addition to values of FA, MD, RD, and AD at each point along the maximum a posteriori path, pathstats.byvoxel.txt now also contains the mean values of the same measures over the entire cross-section of the path distribution at each point along the trajectory of the path.
NEW in this version: Longitudinal tractography. Note: This is an undocumented, unpublished, and only mildly tested BETA provided for testing purposes, so proceed at your own risk. All you need to do to use it is set the baselist variable in your configuration file, see $FREESURFER_HOME/bin/dmrirc.long.example. It assumes that the longitudinal FreeSurfer stream has been run on the anatomical data to obtain the base templates.
TRACULA in FreeSurfer 5.1:
2012/01/09 -- trac-all: This update for trac-all is recommended for all users of version 5.1.
2011/08/17 -- dmri_5.1_snow_leopard.tar.gz: TRACULA executables compiled on snow leopard. This build resolves memory issues that snow leopard users had with the original leopard build.
2011/05/24 -- First release of TRACULA in FreeSurfer 5.1.
References
If you use TRACULA, please cite:
Automated probabilistic reconstruction of white-matter pathways in health and disease using an atlas of the underlying anatomy. Yendiki A, Panneck P, Srinivasan P, Stevens A, Zöllei L, Augustinack J, Wang R, Salat D, Ehrlich S, Behrens T, Jbabdi S, Gollub R and Fischl B (2011). Front. Neuroinform. 5:23. doi: 10.3389/fninf.2011.00023
Using diffusion MRI data acquired with ultra-high gradients to improve tractography in routine-quality data. Maffei C, Lee C, Planich M, Ramprasad M, Ravi N, Trainor D, Urban Z, Kim M, Jones R, Henin A, Hofmann S, Pizzagalli D, Auerbach R, Gabrieli J, Whitfield-Gabrieli S, Greve D, Haber N, Yendiki A (2021). NeuroImage 245:118706.
If you use our measures of head motion, please cite:
Spurious group differences due to head motion in a diffusion MRI study. Yendiki A, Koldewyn K, Kakunoori S, Kanwisher N, Fischl B (2014). NeuroImage 88:79–90.
If you use the longitudinal stream of TRACULA, please cite:
Joint reconstruction of white-matter pathways from longitudinal diffusion MRI data with anatomical priors. Yendiki A, Reuter, M, Wilkens P, Rosas H D, Fischl B (2016). NeuroImage 127:277–286.
