Brodmann Area Maps (BA Maps) and Hinds V1 Atlas
The Martinos Center for Biomedical Imaging and the Institute of Neurosciences and Biophysics (INB) have created an atlas of a subset of the Brodmann Areas, for usage in Freesurfer. The subject set consists of 10 neurologicially normal subjects.
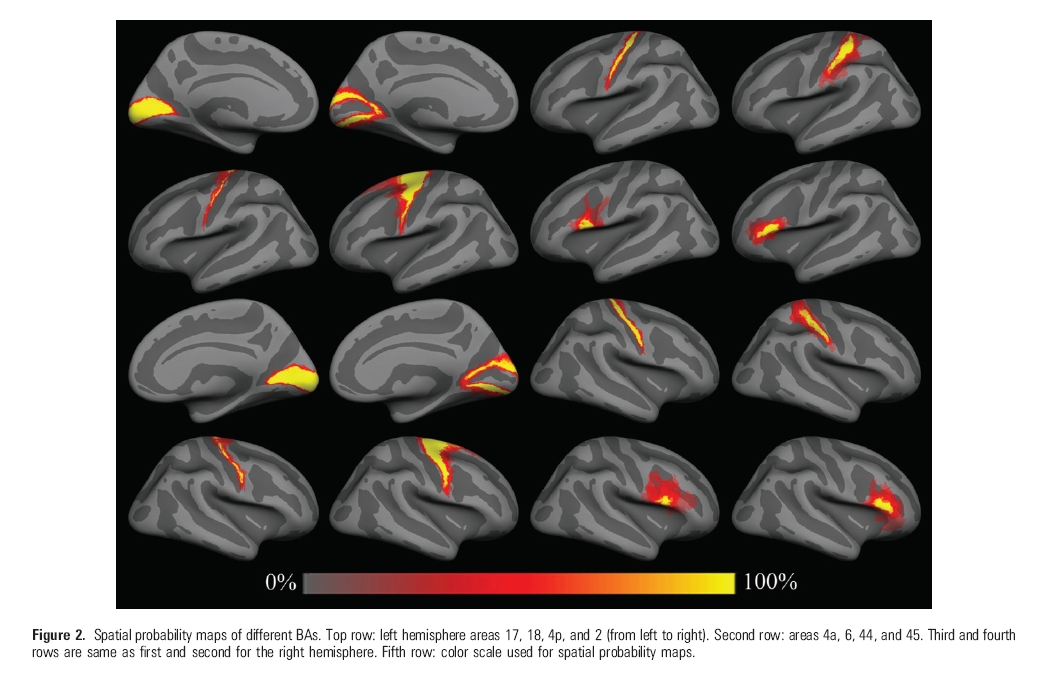
Note: maps are for neuroscientific purposes only! (no commercial use)
BA Labels
The following labels are included in Freesurfer v4.0.3 (and later) for display on the 'fsaverage' surface distributed with Freesurfer:
BA 1 - somatosensory area
BA 2 - somatosensory area
BA 3a - somatosensory area
BA 3b - somatosensory area
BA 4a - primary motor area (anterior)
BA 4p - primary motor area (posterier)
BA 6 - pre-motor area
BA 44 - Broca's area (pars opercularis)
BA 45 - Broca's area (pars triangularis)
V1 (BA 17) - primary visual area
V2 (BA 18) - secondary visual area
V5/MT - visual area, middle temporal
These labels are created by default when recon-all is run. They can be created manually by running:
recon-all -s subjid -ba-labels
To view a label, open an inflated surface (tksurfer subjid lh inflated) and then select File->Label->Load Label and select a label (lh.BA1_exvivo.label, for example). Then select Tools->Labels->Copy Label Statistics to Overlay. Then select View->Configure->Overlay and set Min threshold to 0 and Max threshold to 0.5, and also select Linear threshold, and then press Apply and Close. The label should appear on the surface (you will probably need to press the Redraw button). Only one label at a time can be displayed. The labels show the frequency estimates of the probability that each (displayed) point was part of that BA.
See also Entorhinal and Perirhinal.
V1 Atlas
See the O. Hinds paper 'Accurate prediction of V1 location from cortical folds in a surface coordinate system', in References below, for details. This is different set of work from the Brodmann labels just described, but is mentioned here because it is an alternative V1 label.
The Hinds V1 labels are produced by running:
recon-all -s subjid -label_v1
This will output:
<subjid>/label/?h.v1.prob.label - contains stat values
<subjid>/label/?h.v1.predict.label - thresholded
Note: This requires the subject found in $FREESURFER_HOME/subjects/V1_average to be located (symlinked or copied) in your $SUBJECTS_DIR.
References
Cortical Folding Patterns and Predicting Cytoarchitecture, Fischl, B., N. Rajendran, E. Busa, J. Augustinack, O. Hinds, B.T.T. Yeo, H. Mohlberg, K. Amunts, K. Zilles, (2008). Cerebral Cortex 2008, doi:10.1093/cercor/bhm225.
Accurate prediction of V1 location from cortical folds in a surface coordinate system, Hinds, O.P., N. Rajendran, J.R. Polimeni, J.C. Augustinack, G. Wiggins, L.L. Wald, H.D. Rosas, A. Potthast, E.L. Schwartz and B. Fischl (2007). NeuroImage 2007.10.033.
- Quantitative analysis of cyto- and receptor architecture of the human brain, Zilles K, Schleicher A, Palomero-Gallagher N, Amunts K (2002). Brain Mapping: The Methods, edited by J. C. Mazziotta and A. Toga, USA: Elsevier, 2002, p. 573-602.
Cytoarchitecture of the cerebral cortex - more than localization, Amunts K, Schleicher A, Zilles K (2007). NeuroImage 37:1061-1065.
- Towards multimodal atlases of the human brain, Toga AW, Thompson PM, Mori S, Amunts K, Zilles K (2006). Nature Neuroscience Reviews, 7(12):952-966.
- Gender-specific left-right asymmetries in human visual cortex, Amunts K, Armstrong E, Malikovic A, Hoemke L, Mohlberg H, Schleicher A, Zilles K (2007). The Journal of Neuroscience, 27(6):1356-1364.
