Introduction to FreeSurfer Output DRAFT
Introduction to FreeSurfer Output
Exercise 1
Difficulty: Beginner
Goal: Practice basic Freeview tasks.
In the examples above you looked at data from a subject called “good_output”. For this challenge complete the following tasks for subject “004”.
- Open the subject’s aparc+aseg.mgz volume with a colormap of “lut”.
- Swap the view to sagittal
- Navigate with the arrow keys to find the right putamen
Need a hint?
Here is how you opened up a similar volume for the “good_output” subject: freeview -v good_output/mri/aseg.mgz:colormap=lut
Please note that we were in the $TUTORIAL_DATA/buckner_data/tutorial_subjs directory when we used that command - so cd there if you'd like to base your command off the above example.
Want to know the answer? Click and drag to highlight and reveal the text below.
cd $TUTORIAL_DATA/buckner_data/tutorial_subjs |
freeview -v 004/mri/aparc+aseg.mgz:colormap=lut |
Exercise 2
Difficulty: Beginner
Goal: Practice visualizing data with overlays.
- Open 004’s lh.pial surface, with the overlay named lh.thickness, set the overlay to display with a threshold of 1,2
- Look up vertex 141813
- What is the thickness and label of this vertex?
Need a hint?
Here is how you opened up a similar surface for the good_output subject, with thickness information: freeview -f good_output/surf/lh.inflated:overlay=lh.thickness:overlay_threshold=0.1,3 --viewport 3d
And here is how you opened up a similar surface for the good_output subject with the Desikan-Killany parcellation: freeview -f good_output/surf/lh.pial:annot=aparc.annot
Please note that we were in the $TUTORIAL_DATA/buckner_data/tutorial_subjs directory when we used that command - so cd there if you'd like to base your command off the above example.
Want to know the answer? Click and drag to highlight and reveal the text below.
cd $TUTORIAL_DATA/buckner_data/tutorial_subjs |
freeview -f 004/surf/lh.pial:overlay=lh.thickness:overlay_threshold=1,2 --viewport 3d |
Exercise 3
Difficulty: Beginner
Goal: Practice opening multiple files at a time with FreeView.
- For this challenge start with this terminal command:
freeview -v 004/mri/wm.mgz:colormap=jet 004/mri/brainmask.mgz -f 004/surf/lh.pial:edgecolor=blue 004/surf/lh.white:edgecolor=red
- Right now it only opens the left hemisphere pial and white matter surfaces, alter it to open both for the right hemisphere as well, with colors that match the left hemisphere side.
- Once you have the last command working, rearrange the volume layers in freesurfer so that the wm.mgz is at %20 opacity and the brainmask can be seen underneath it (this can also be done through altering the terminal command - you can choose which way to do so).
- You will need to be in $TUTORIAL_DATA/buckner_data/tutorial_subjs for the command to work, so cd there if needed
Want the solution? Click and drag to highlight and reveal the text below.
cd $TUTORIAL_DATA/buckner_data/tutorial_subjs |
freeview -v 004/mri/brainmask.mgz 004/mri/wm.mgz:colormap=jet:opacity=.2 -f 004/surf/lh.pial:edgecolor=blue 004/surf/lh.white:edgecolor=red 004/surf/rh.pial:edgecolor=blue 004/surf/rh.white:edgecolor=red |
Exercise 4
Difficulty: Intermediate - assumes some comfort with navigating Unix and FreeView
Goal: Export a series of images from FreeView and stitch them together to create a GIF.
- Open up any surface from the tutorial data
Set the viewport to 3d view, right click in the viewport and select Hide All Slices
In the File menu, select Save Movie Frames
Set up the options as in the following picture - you will likely want to create a new directory in your home directory to save the output to.
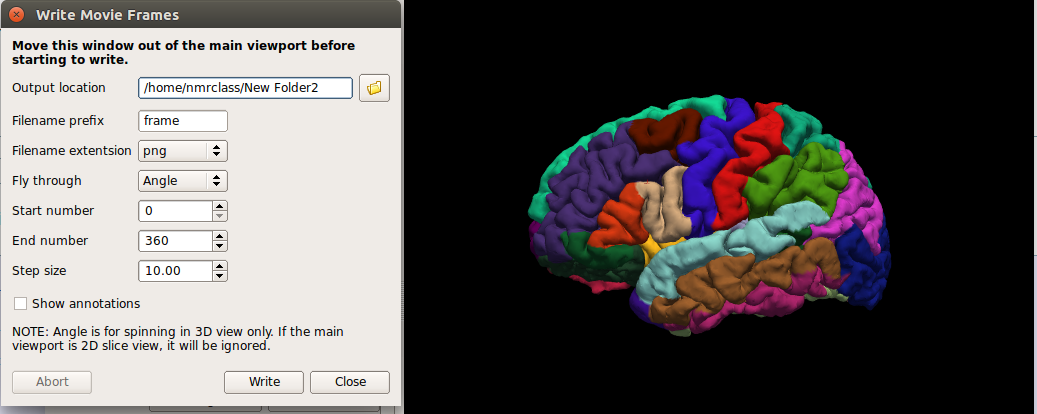
- In a terminal, navigate to the new directory you output the movie data to.
Run this command: convert -delay .1 *.png brainanim.gif
Note: convert is from the ImageMagick library, which is a prerequesite for running FreeView.
-delay determines the gap between frames, *.png selects all png files in the working directory, brainanim.gif is the output name.
To view your GIF, open it with firefox firefox brainanim.gif
Practice Working With Data
Exercise 1
Difficulty: Beginner
Goal: Prepare a dicom series for the recon-all stream
Your goal is to set up your environment variables and assemble the correct recon-all command to process a dicom series.
To begin navigate to the following directory in your terminal: $TUTORIAL_DATA/practice_with_dicoms
Take a look at the directories found there. The dicoms directory contains a dicom series, and the subjects directory is where your recon-all subject output should go. You will want to use dcmunpack to find the first image of the first T1w_MPR_vNav_4eRMS series, set your SUBJECT_DIR environment variable, then then type out and run your recon-all command (use practice_subject as the subject name). Make sure the command starts without any errors - and if it does cancel the process by pressing ctrl and c on the keyboard (a recon-all can take many hours!).
Check in the work directory to ensure a directory with your subject's name was created (practice_subject), if you see the folder you have completed the challenge!
Hints:
- Follow the tutorial example for guidance
- It might be tricky to find the path to the subjects directory if you are new to Unix, you can use this: $TUTORIAL_DATA/practice_with_dicoms/subjects
Want to see the answer? Highlight the lines below
cd $TUTORIAL_DATA/practice_with_dicoms \ |
cd dicoms \ |
dcmunpack -src . -scanonly scan.log |
export SUBJECTS_DIR = $TUTORIAL_DATA/practice_with_dicoms |
recon-all -i MR.1.3.12.2.1107.5.2.43.67026.2019072908432986436303794 -s practice_subject |
Anatomical ROI Analysis
Exercise 1
Difficulty: Beginner
Goal: To practice collecting different types of measures with asegstats2table
Create a table called mean.practice.table that lists the average mean intensities of all segments for subjects 004 021 and 092.
When done use the following command to open up an excel-like program on your computer and look at the data soffice --calc mean.practice.table , note, the command may take some time to run and may report warnings which you can ignore. (If you are not at a FreeSurfer course, you may not have this program, in this case use gedit mean.practice.table to open the table. )
Hints:
- If you don't specify which segment numbers you want, the measurements will be collected for all segments.
If you run asegstats2table --help you can get a list of all the ways to configure your table, here is some information from that command which might help:
--meas=MEAS measure: default is volume ( alt: mean, std)
For example, asegstats2table --meas std would add the standard deviation measurements of each segment to the table.
You will probably want to run cd $SUBJECTS_DIR first so you are in the right directory.
Want the answer? Highlight the black lines below to see!
cd $SUBJECTS_DIR \ |
asegstats2table --subjects 004 021 092 --meas mean --tablefile mean.practice.table / |
less mean.practice.table |
Exercise 2
Difficulty: Intermediate, will present challenges to Unix beginners (but is totally possible!)
Goal: To practice collecting different types of measures and using different atlases with aparcstats2table
Create a table called rh.aparc.a2009.thickness.table which lists the main thickness in all left hemisphere cortical parcellations for subjects 004 021 and 040.
When done use the following command to open up an excel-like program on your computer and look at the data soffice --calc lh.aparc.a2009.thickness.table , note, the command may take some time to run and may report warnings which you can ignore. (If you are not at a FreeSurfer course, you may not have this program, in this case use gedit mean.practice.table to open the table. )
Hints:
If you run aparcstats2table --help you can see a list of all the different ways to configure your table, here is some information found through that command that might help:
-p PARC, --parc=PARC parcellation.. default is aparc ( alt aparc.a2009s)
-m MEAS, --measure=MEAS measure: default is area ( alt volume, thickness, thicknessstd, meancurv, gauscurv, foldind, curvind)
You will probably want to run cd $SUBJECTS_DIR first so you are in the right directory.
Want the answer? Highlight the black lines below to see!
cd $SUBJECTS_DIR \ |
aparcstats2table --subjects 004 021 040 --hemi lh --meas thickness --parc aparc.a2009s --tablefile lh.aparc.a2009s.thickness.table / |
less mean.practice.table |
soffice --calc lh.aparc.a2009s.thickness.table |
Surface Based Group Analysis
Exercise 1
Difficulty: Beginner
Goal: To practice setting up an analysis on subject surfaces to test an experimental hypothesis.
In the tutorial you just completed you examined the effect of age on cortical thickness, accounting for the effects of gender. In this challenge you will use the same data set to examine the effect of gender on cortical thickness, accounting for the effects of age.
Your goal is to create a new contrast file and alter the commands you used above to examine this new hypothesis. You will then open the new sig.mgh overlay which will highlight significant differences found.
Please name your new contrast file challenge-Cor.mtx Please set your GLM output directory to glm_challenge
Hints:
It will help if you start in this directory $SUBJECTS_DIR/glm and run all of your commands from there.
You will want to use mkdir glm_challenge to make the output directory, and be sure to use that directory when you run mri_glmfit
- Follow the tutorial for guidance.
- You will not need to change the FSGD file, as you are using the same groups - you don't need to change the descriptor of the groups.
You can use the command touch challenge-Cor.mtx to create a new contrast, and gedit challenge-Cor.mtx to edit it.
You will need to adjust your contrasts, consider the following snippet from the wiki page Two Groups (One Factor/Two Levels), One Covariate
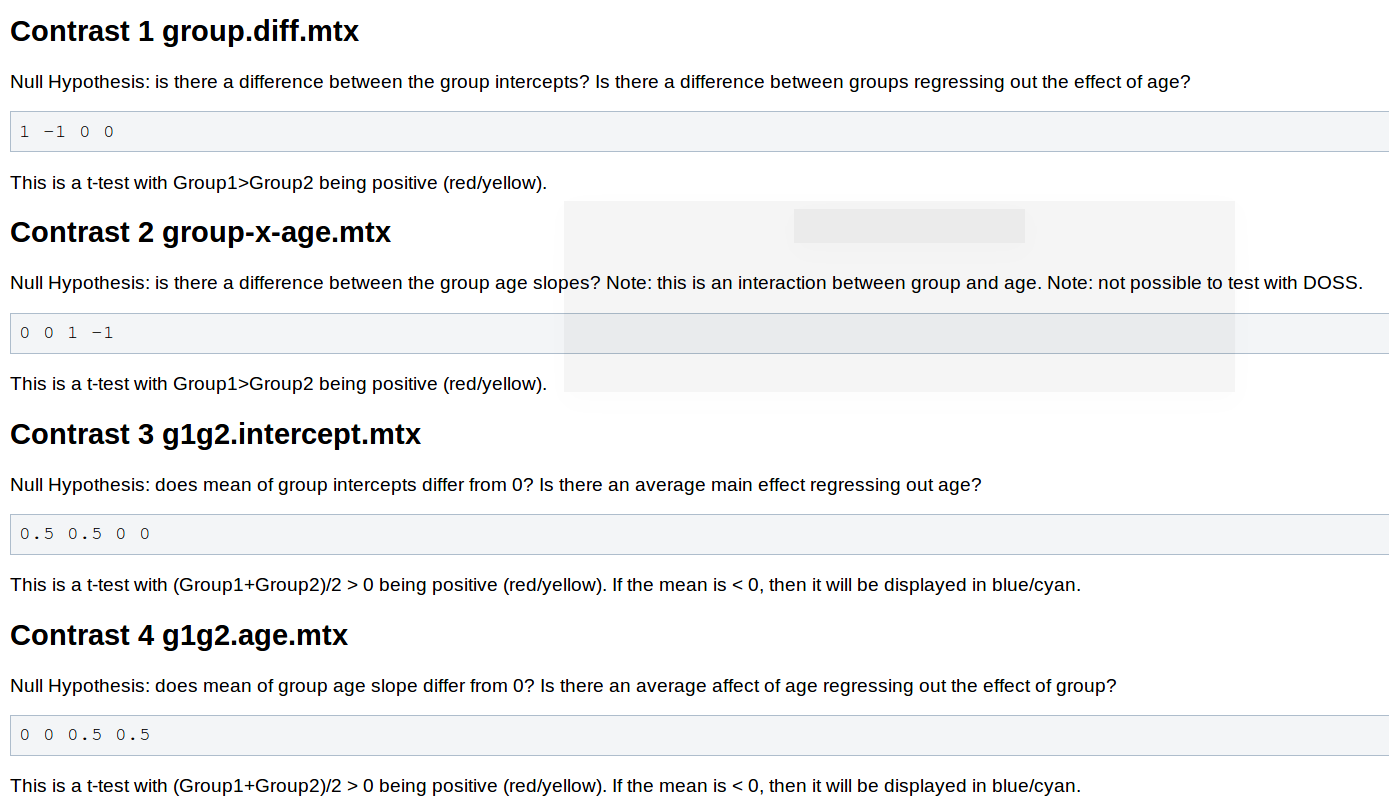
- You will not need to pre-process the data, as changing the contrast doesn't have any effect on this stage.
- When you run mri_glmfit you will want to change which contrast it reads and the directory it saves to.
If you named your contrast challenge-Cor.mtx and set your output directory to glm_challenge than this command should let you examine your significance overlay.
freeview -f $SUBJECTS_DIR/fsaverage/surf/lh.inflated:overlay=glm_challenge/challenge-Cor/sig.mgh:overlay_threshold=4,5 -viewport 3d -layout 1
Want to know the answer? Highlight the text below
cd $SUBJECTS_DIR/glm |
touch challenge-Cor.mtx |
gedit challenge-Cor.mtx |
contrasts should be 1 -1 0 0 |
mri_glmfit --y lh.gender_age.thickness.10.mgh --fsgd gender_age.fsgd dods --C challenge-Cor.mtx --surf fsaverage lh --cortex --glmdir glm_challenge |
freeview -f $SUBJECTS_DIR/fsaverage/surf/lh.inflated:overlay=glm_challenge/challenge-Cor/sig.mgh:overlay_threshold=4,5 -viewport 3d -layout 1 |
You should see a few very small areas of significance, in the next tutorial you will see how running permutations affects this analysis | |
Clusterwise Correction For Multiple Comparisons (Permutations)
Exercise 1
Difficulty: Intermediate
Goal: Practice running permutation simulations to correct for multiple comparisons.
Your goal is to run the same permutation operations you learned about in this tutorial on the analysis you created from last tutorial ( where you examined the effect of gender on cortical thickness). In this challenge you will will actually run the permutations (in the tutorial above it was done for you) - but you will do it only run it with 10 rather than the usual 1000 to save time. When done open up the same overlays as you did in the tutorial and see what effect the smoothing did on the analysis.
Hints:
- Follow the tutorial for guidance
- You will want to start working in the directory from last tutorial (because that is where your data is):
export SUBJECTS_DIR=$TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial
cd $SUBJECTS_DIR/glm
You will need to run mri_glmfit again with the --eres-save option, be sure to run it with the contrast and directory you made from before ( challenge-Cor.mtx and glm_challenge )
When you run the simulation - be sure to change the directory and the permutations count to 10 --perm 10 4.0 abs
- If you followed the file and directory names specified in the challenge, this command should let you view your output:
freeview -f $SUBJECTS_DIR/fsaverage/surf/lh.inflated:overlay=glm_challenge/challenge-Cor/perm.th40.abs.sig.cluster.mgh:overlay_threshold=2,5:annot=glm_challenge/challenge-Cor/perm.th40.abs.sig.ocn.annot -viewport 3d -layout 1
Want to know the answer? Highlight below to find out.
cd $SUBJECTS_DIR/glm |
mri_glmfit --y lh.gender_age.thickness.10.mgh --fsgd gender_age.fsgd dods --C challenge-Cor.mtx --surf fsaverage lh --cortex --glmdir glm_challenge --eres-save |
mri_glmfit-sim --glmdir glm_challenge --perm 10 4.0 abs --cwp 0.05 --2spaces --bg 1 |
freeview -f $SUBJECTS_DIR/fsaverage/surf/lh.inflated:overlay=glm_challenge/challenge-Cor/perm.th40.abs.sig.cluster.mgh:overlay_threshold=2,5:annot=glm_challenge/challenge-Cor/perm.th40.abs.sig.ocn.annot -viewport 3d -layout 1 |
Registration
Exercise 1
Difficulty: Intermediate
Goal: Practice using bb register to automatically register unaligned data.
Navigate to $SUBJECTS_DIR/multimodal/fmri/challenge/challenge-template , which contains template.nii which is a volume you need to align to a subject named regsubject . regsubject is in the subjects directory that you have already set if you have been following the tutorial above. Name your output for bbregister register.lta
You can use the command freeview -v template.nii $SUBJECTS_DIR/regsubject/mri/orig.mgz:visible=0 -f $SUBJECTS_DIR/regsubject/surf/lh.white $SUBJECTS_DIR/regsubject/surf/rh.white to inspect the unaligned data first if you would like.
When done, inspect the data to see if it is aligned. Note - the results may be slightly unexpected! If you names your output as requested above the following command should open everything up:
freeview -v $SUBJECTS_DIR/regsubject/mri/orig.mgz:visible=0 template.nii:reg=register.lta -f $SUBJECTS_DIR/regsubject/surf/lh.white $SUBJECTS_DIR/regsubject/surf/rh.white
Want the answer? Highlight below to see:
cd $SUBJECTS_DIR/multimodal/fmri/challenge/challenge-template |
bbregister --mov template.nii --bold --s regsubject --lta register.lta |
