This process provides the user with flattened images of the whole brain or select parts (e.g. occipital lobe) of the brain.
Cutting the Full Surface
The full surface represents an entire hemisphere without the mid-brain (middle of the medial surface). First, load the subject surface into tksurfer:
tksurfer subjid lh inflated
Load the curvature through the File->Curvature->Load menu (load lh.curv). This should show a green/red curvature pattern. Red = sulci.
Right click before making a cut; this will clear previous points. This is needed because it will string together all the previous places you have clicked to make the cut.
- Make 5 relaxation cuts. Note that these cuts extend from the medial wall to the outer edge where the lateral surface would begin. There are no cuts on the lateral surface. For each cut, select the points (left-click) and then press the Cut icon (scissors with an open triangle for a line cut). Make the points fairly close together; if they are too far apart, the cut fails.
- 1 cut down the calcarine (same cut as for the occiput surface)
- 3 equally spaced radial cuts on the medial surface
- 1 sagitally oriented cut around the temporal pole
- Encircle midline region (corpus callosum and mid-brain structures) to remove:
- Left-click a circle of points, being sure to encompass the ends of the radial cuts
- Once the circle of points are selected, click the Cut Closed Line icon
- Single left-click in the newly cut circle
- Press Fill Uncut Area button (icon = filled triangle)
- Save patch:
Select File->Patch->Save Patch As. Specify the patch name in the Selection field:
- The default names for the full cortical surfaces with the midline removed are:
- lh.full.patch.3d
- rh.full.patch.3d
- The default names for the full cortical surfaces with the midline removed are:
- Create flattened patch:
mris_flatten -w 10 lh.full.patch.3d lh.full.flat.patch.3d
The -w 10 option writes the current state of the flattening every 10 iterations of the algorithm. This allows observing the flattening, as the algorithm will take more than hours to complete, and it's helpful to have some preliminary images after a few minutes. The filename written is shown in the output. In tksurfer, use File->Patch->Load Patch to load that flattened image (you will probably have to zoom-out and rotate to see it).

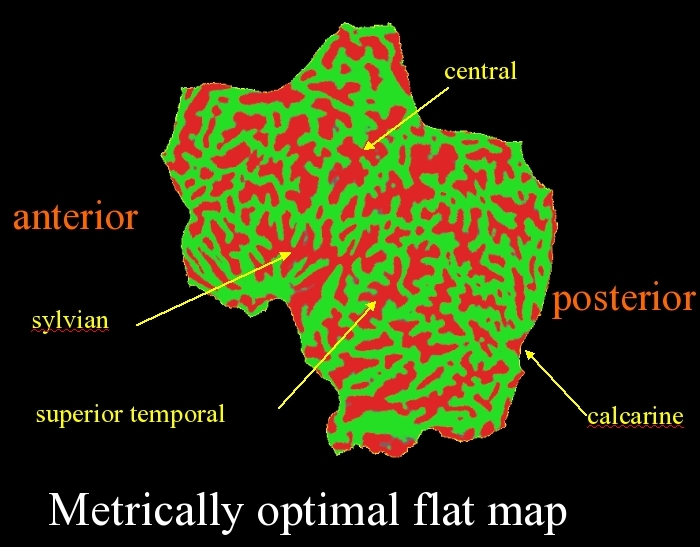
Cutting the Occiput Surface
The occiput patch represents the occipital pole with a cut down the calcarine fissure. This cortical patch is particularly useful for displaying the results of visual experiments.
In tksurfer:
- Make calcarine sulcus relaxation cut.
- Select points (left-click) along the calcarine fissure beginning at the occipital pole and moving anteriorly.
- Open-line cut with CUTLINE (surfer tools).
- Specify cutting plane.
- Select three points (labeled 1, 2, and 3 below) to define the cutting plane, and a fourth point (labeled 4 below) to specify which portion of the surface to keep (left-click).
- Make the planar cut.
- CUTPLANE button.
- Specify the patch name in the patch field and press WRITE.
- ...
- The default names for the occiput cortical surfaces are:
- rh.occip.patch.3d
- lh.occip.patch.3d
