Troubleshooting your output
This set of exercises will take you through a few examples of problem outputs, asking you to identify the problems and possible methods to fix the problems. Each example will have a before and after picture, as well as an explanation of how to fix the problems seen.
Preparations
If You're at an Organized Course
If you are taking one of the formally organized courses, everything has been set up for you on the provided laptop. The only thing you will need to do is run the following commands in every new terminal window (aka shell) you open throughout this tutorial. Copy and paste the commands below to get started:
setenv SUBJECTS_DIR $TUTORIAL_DATA/buckner_data/tutorial_subjs cd $SUBJECTS_DIR
To copy: Highlight the command in the box above, right click and select copy (or use keyboard shortcut Ctrl+c), then use the middle button of your mouse to click inside the terminal window (this will paste the command). Press enter to run the command.
These two commands set the SUBJECTS_DIR variable to the directory where the data is stored and then navigates into this directory. You can now skip ahead to the tutorial (below the gray line).
If You're not at an Organized Course
If you are NOT taking one of the formally organized courses, then to follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names. These are the commands that you need to run before getting started:
tcsh source your_freesurfer_dir/SetUpFreeSurfer.csh setenv SUBJECTS_DIR $TUTORIAL_DATA/buckner_data/tutorial_subjs cd $SUBJECTS_DIR
Notice the command to open tcsh. If you are already running the tcsh command shell, then the 'tcsh' command is not necessary. If you are not using the tutorial data you should set your SUBJECTS_DIR to the directory in which the recon(s) of the subject(s) you will use for this tutorial are located.
Failure Modes
Below, we ask you to look at several subjects with various errors that require manual intervention. Open each subject and try to identify the problem on your own. For each subject, there is a link to instructions on how to fix the problem. While you are trying to identify the problems with the subjects listed below you may find it helpful to refer back to the previous exercise, or open your own instance of the subject good_output to compare these subjects to a good example.
Edits to the wm volume
Now, take a look at the next subject, wm1_edits_before.
freeview -v wm1_edits_before/mri/wm.mgz:colormap=heat:opacity=0.4 wm1_edits_before/mri/brainmask.mgz \ -f wm1_edits_before/surf/lh.white:edgecolor=blue wm1_edits_before/surf/lh.pial:edgecolor=red \ wm1_edits_before/surf/rh.white:edgecolor=blue wm1_edits_before/surf/rh.pial:edgecolor=red
Again, this will bring up the brainmask.mgz volume, the wm.mgz volume, and the surfaces for both hemispheres.
Click in the main viewing area and use Alt-C or the arrows  to flip between the wm.mgz and brainmask.mgz on top of each other.
to flip between the wm.mgz and brainmask.mgz on top of each other.
with the wm.mgz loaded on top of the brainmask.mgz volume, it should look like this: 
In your second and third terminal window, if not already open, open the surfaces in tksurfer:
tksurfer wm1_edits_before lh inflated
tksurfer wm1_edits_before rh inflated
This will open the inflated surfaces for both hemispheres in tksurfer.
The trouble with this subject has occurred during the white matter segmentation step. Check the surfaces in the T1 volume to find the spot that does not match the actual gray/white boundary. If you load the wm.mgz volume as the aux volume you will see a hole in this area. If you are looking in tksurfer you will see a hole or dimple in the inflated surface. You can use the "save point" "goto point" commands to find this spot in the volume.
Using the Page Up and Page Down keys, you can scroll towards the anterior portion of the brain until you find an area in the right hemisphere where the white surface (blue line) does not follow the surface of the brain, but in fact cuts into it. This geometric innacuracy is cause by a lesion where the white matter has been marked as non-white matter. 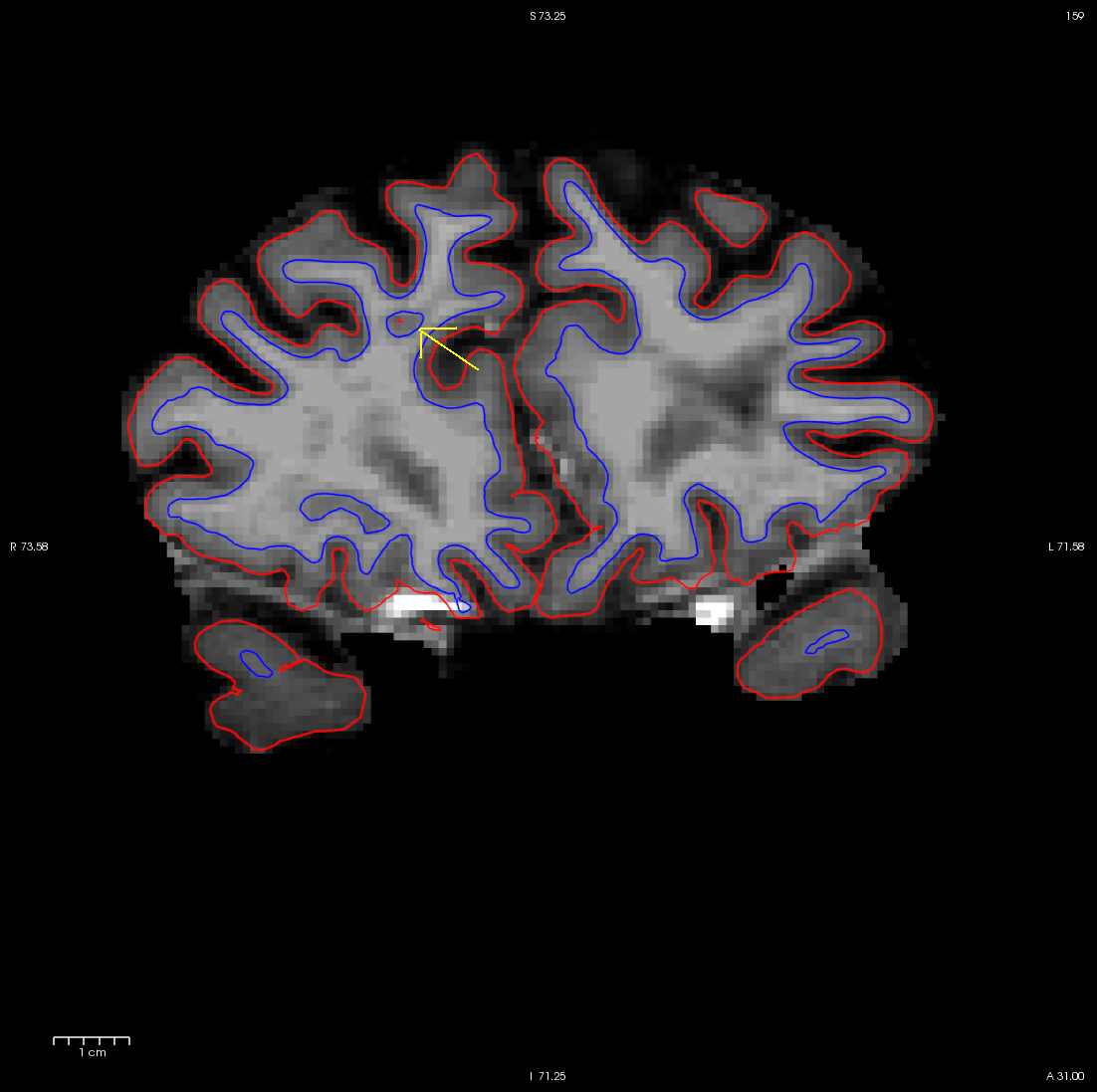
use Alt-C to have the wm.mgz volume loaded on top and you can see that this area has been left out of the wm volume completely.
You can see this as a dimple or hole on the inflated surface in tksurfer too (see next image). If not already open, in a second terminal window, open the rh surface in tksurfer and you can see the dimple or hole on the medial side:
tksurfer wm1_edits_before rh inflated
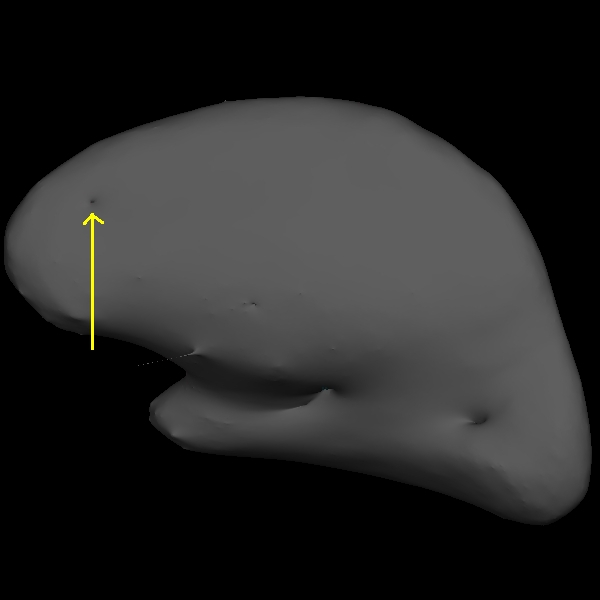
To fix this problem you will need to fill in the missing voxels in the wm.mgz volume. First, it's a good idea to use the page up and down keys to scroll through the individual slices in the volume until you get a good idea of where the problem starts and ends. You will want to start filling in voxels when the inaccuracy appears, and keep filling them in slice by slice until the problem is no longer visible. The coronal view and a brush radius of one or two are good settings for painting in voxels. Zoom In and Out either by using the scroll button on your mouse or holding down shift and the up or down keys.
To begin editing voxels, make sure the wm.mgz is highlighted in the volume menu, and on top on the brainmask.mgz. click on the edit voxels option (  ) and enter 255 into the brush value box. your menu should look like this:
) and enter 255 into the brush value box. your menu should look like this:
Remember: the wm.mgz must be on top and and the brush value 255 in order to edit voxels in the wm.mgz, if it is not visible on top you will be editing the brainmask.mgz. You should only be editing the brainmask for pial edits and not wm edits.
In the Voxel edit menu that pops up, click on the first icon "freehand" and select a Brush size of 1 or 2. Make sure the Reference is the wm volume. This menu should look like this:
You are now ready to edit. click back on the volume, and using your left mouse button fill in the wm hole with voxels. If you fill in too many, you can press shift while using your left mouse button to erase voxels. You can keep flipping back between the brainmask and the wm volume using your Alt-C combination if you want to check that what you are filling in is indeed white matter. At any time you can use the CTRL-Z combination to undo as many actions as you like. When you're done editing the volume should look like this.
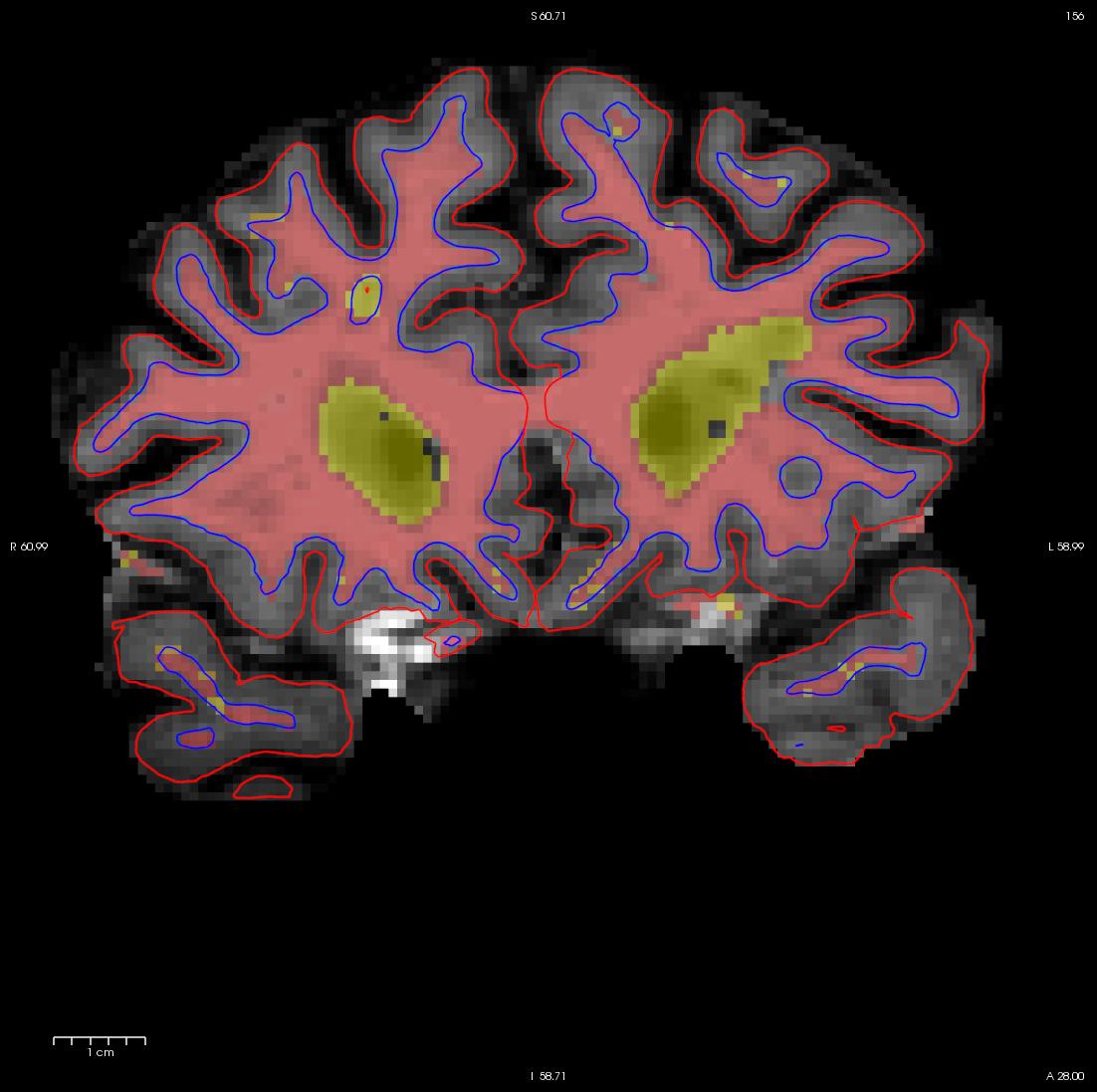
While filling the hole in slice 160, you will notice that the hole is not completely enclosed. To see where the boundary should be, switch back and forth between the brainmask.mgz and wm.mgz volume with Alt-C. Use the brainmask.mgz volume as a guide, and make the boundary of the voxels that you paint in correspond as closely as possible to the boundaries suggested in the brain volume. Go to the next slice, and fill in this slice in the same way. You will need to continue filling in slices in this fashion until the region is completely filled in. If you would like to compare your edits to ones that have been done for you you can open a second freeview window with the wm1_edits_after wm.mgz volume, and for each slice check your results with the corresponding slice in the wm1_edits_after volume. At any time, you can save the changes you've made by clicking on the wm.mgz, making sure it is highlighted in the menu to the left side of the viewing area and click the save volume button  or save in the file menu. You can verify your results by viewing the changes made to the wm1_edits_after wm.mgz volume
or save in the file menu. You can verify your results by viewing the changes made to the wm1_edits_after wm.mgz volume
To view this corrected subject (which was corrected by following the detailed instructions provided to you):
freeview -v wm1_edits_after/mri/wm.mgz:colormap=heat:opacity=0.4 wm1_edits_after/mri/brainmask.mgz \ -f wm1_edits_after/surf/lh.white:edgecolor=blue wm1_edits_after/surf/lh.pial:edgecolor=red \ wm1_edits_after/surf/rh.white:edgecolor=blue wm1_edits_after/surf/rh.pial:edgecolor=red
Tips:
- Sometimes regions may appear enclosed in some slices (i.e. appear as holes), open in subsequent slices (i.e. no longer appear as holes), then enclosed again as you scroll forward. The rule of thumb when editing these is to keep filling until you reach the slice where they finally open up and are no longer enclosed.
- When editing geometric inaccuracies in the future, you may find it easier to edit the first slice and last slice first, thereby 'capping' the slices that need filling. Then you just need to fill all the slices between the endcaps.
- Sometimes there is variation between a subject's left and right hemispheres, so that in a particular slice one hemisphere's region will finally 'open up', but the other hemisphere's region is still enclosed. In such cases, continue to fill the enclosed region only, even though the other is open. This will address the topological problem.
- Problems like the one shown here, due to brain lesions, should always be fixed with edits to the wm.mgz volume. Do not use control points to try and automatically adjust the intensity in these areas.
After you have saved all of your edits, you could recreate the final surfaces with the command:
recon-all -autorecon2-wm -subjid wm1_edits_before
Do not run this command if you are conducting the tutorial! This step will take a long time and there is no need to run it for the tutorial purposes.
Non-white matter classified as white matter
Sometimes non-white matter (i.e., skull) is included as white matter. To load in the example use this command:
freeview -v wm2_edits_before/mri/wm.mgz:colormap=heat:opacity=0.4 wm2_edits_before/mri/brainmask.mgz \ -f wm2_edits_before/surf/lh.white:edgecolor=blue wm2_edits_before/surf/lh.pial:edgecolor=red \ wm2_edits_before/surf/rh.white:edgecolor=blue wm2_edits_before/surf/rh.pial:edgecolor=red
Scroll through the brainmask.mgz volume slices and as you go posterior you will see a group of voxels included as white matter, around slice 150. They are surrounded by the white surface (blue line). This group lies outside of the pial surface (red line) and is clearly not white matter. In order to correct this inaccuracy it is necessary to delete the voxels which are not part of the white matter.
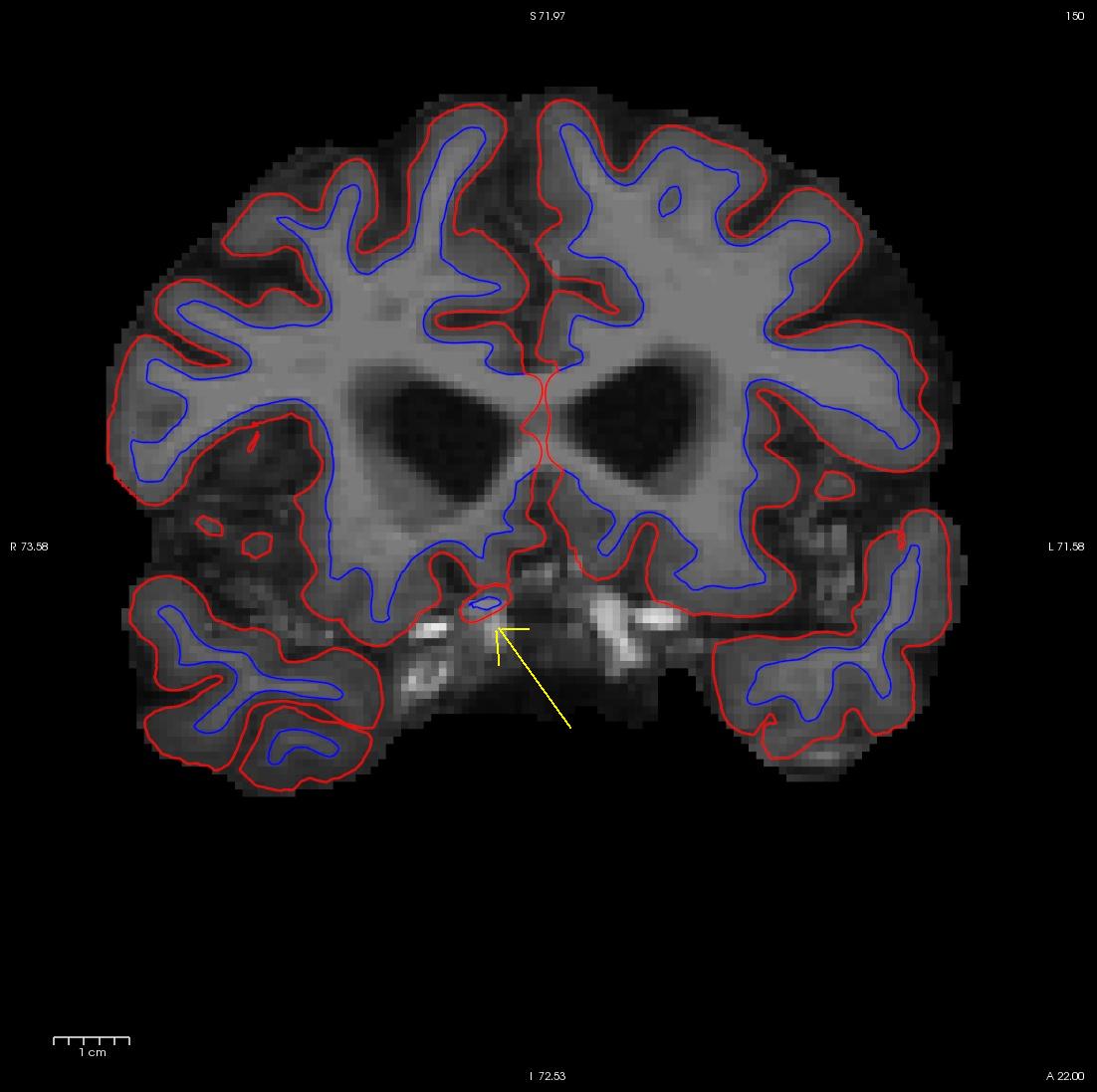
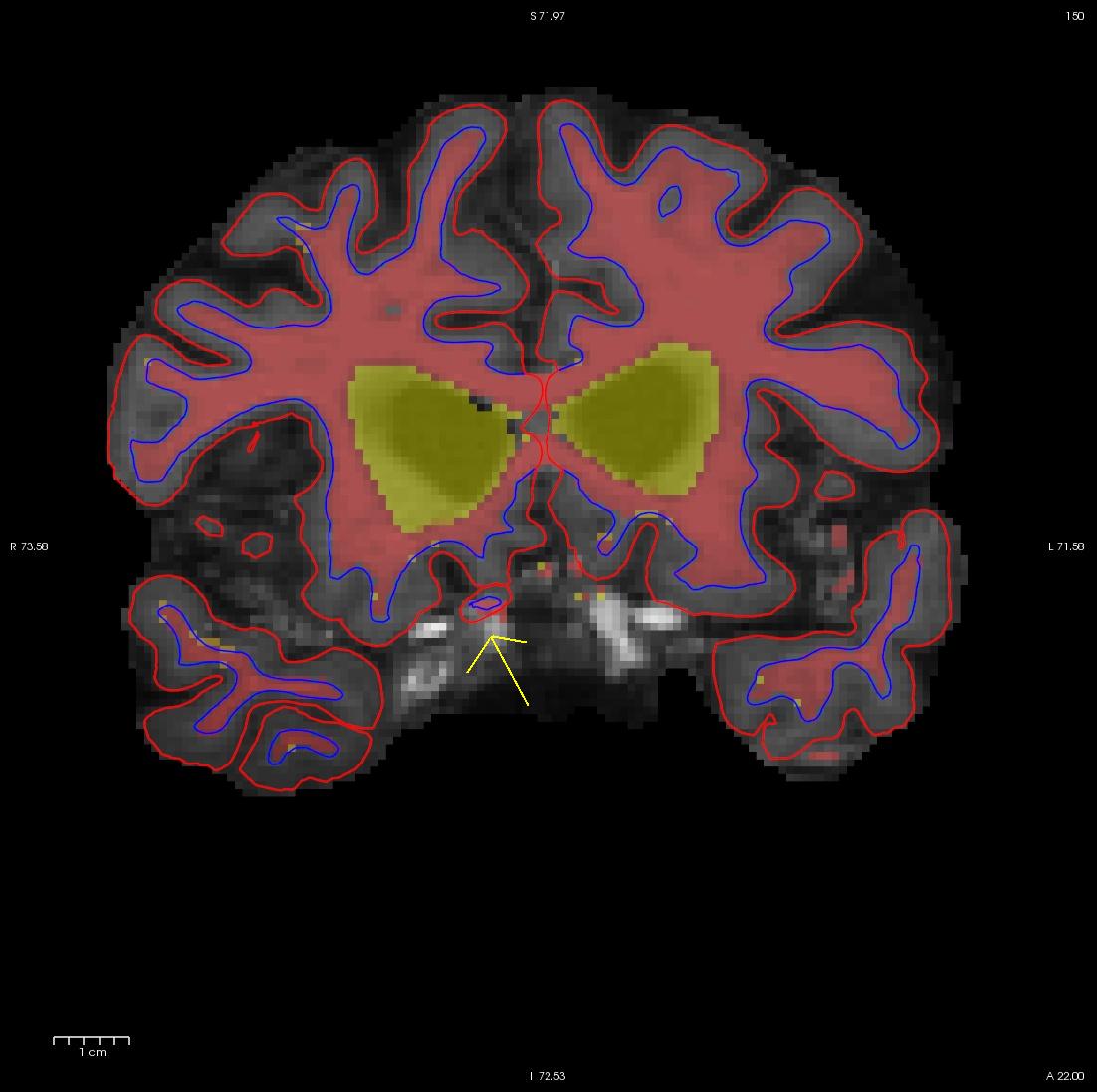
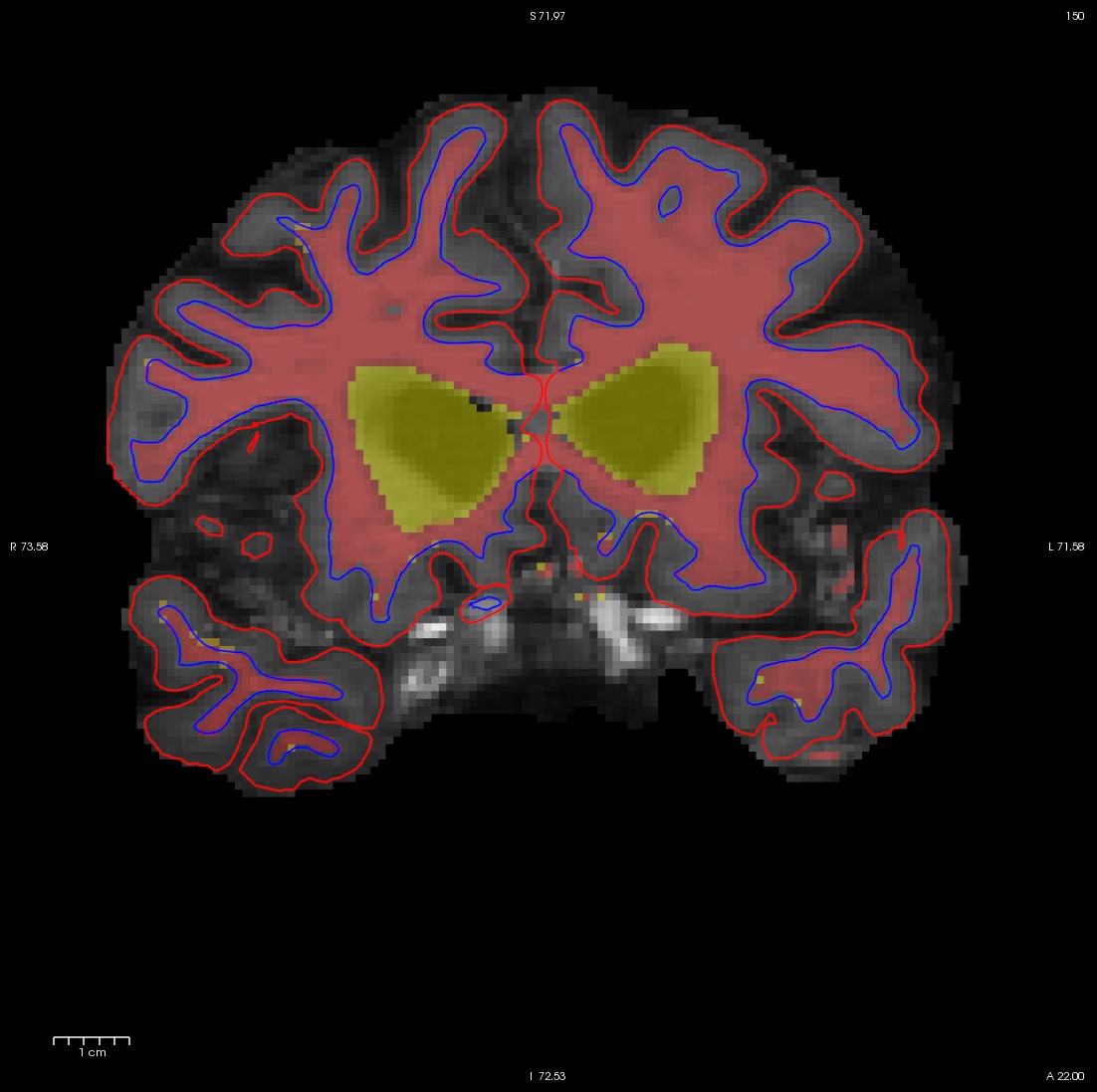
Flipping back and forth between the wm.mgz and the brainmask.mgz you can clearly see that this region should not be labeled as white matter. Click the edit voxels tool again and double check wm as the reference and that the wm.mgz is on top and highlighted. Then hold down your shift key as you deleted the voxels you want. This is what it should look like after you have deleted the errant voxels. When you are done, Click on the wm.mgz, make sure it is highlighted in the menu to the left side of the viewing area and click the save volume button  or save in the file menu.
or save in the file menu.
Once you have made all the edits to the wm.mgz volume you would regenerate the surfaces using the following command:
recon-all -autorecon2-wm -autorecon3 -subjid wm2_edits_before
Do not run this command if you are conducting the tutorial! This step will take a long time and there is no need to run it for the tutorial purposes.
To view this corrected subject (which was corrected by following the detailed instructions provided to you):
freeview -v wm1_edits_after/mri/wm.mgz:colormap=heat:opacity=0.4 wm1_edits_after/mri/brainmask.mgz \ -f wm1_edits_after/surf/lh.white:edgecolor=blue wm1_edits_after/surf/lh.pial:edgecolor=red \ wm1_edits_after/surf/rh.white:edgecolor=blue wm1_edits_after/surf/rh.pial:edgecolor=red
Edits to the brainmask volume
Open the subject, pial_edits_before.
freeview -v pial_edits_before/mri/brainmask.mgz \ -f pial_edits_before/surf/lh.white:edgecolor=yellow pial_edits_before/surf/lh.pial:edgecolor=red \ pial_edits_before/surf/rh.white:edgecolor=yellow pial_edits_before/surf/rh.pial:edgecolor=red
The command above will open the brainmask.mgz volume and the surfaces for both hemispheres. (You can feel free to open other volumes using the load volume button  or to load in the aseg.mgz if you want or need to.
or to load in the aseg.mgz if you want or need to.
If you would like to see the inflated surfaces for each hemisphere of this subject, open a second and third terminal window, then open the surfaces in tksurfer:
tksurfer pial_edits_before lh inflated
tksurfer pial_edits_before rh inflated
Correcting Pial Surfaces
The pial surface is created by expanding the white matter surface so that it closely follows the gray-CSF intensity gradient as found in the brainmask.mgz volume. Once an accurate white surface is created then you can work on correcting the pial surface if needed. The pial surface boundary and white matter surface boundary should not cross. After the pial surface has been generated, it's a good idea to visually check it for defects that may have been created during automatic topology fixing. To check the pial surface, it may be loaded into tkmedit and viewed along with the brainmask.mgz volume. If the surface appears not to follow the gray-CSF boundary in the volume, edits may be required.
Editing the Volume
Subject pial_edits_before is an example of the pial surface including non-cortex within the boundaries. This page will take you through fixing this and other similar problems with the pial surface including non-cortex material.
First, make sure you have pial_edits_before loaded in freeview:
freeview -v pial_edits_before/mri/brainmask.mgz -f pial_edits_before/surf/lh.white:edgecolor=yellow \ pial_edits_before/surf/lh.pial:edgecolor=red pial_edits_before/surf/rh.white:edgecolor=yellow \ pial_edits_before/surf/rh.pial:edgecolor=red
Use the page up and down keys to go through the volume slice by slice, and view the pial surface (red line) and white matter surface (yellow line). Notice the bright diagonal line in slice 157 that has caused the pial surface to expand past the actual pial boundary. This is the result of a bad segmentation incorporating a piece of the dura within the pial surface.
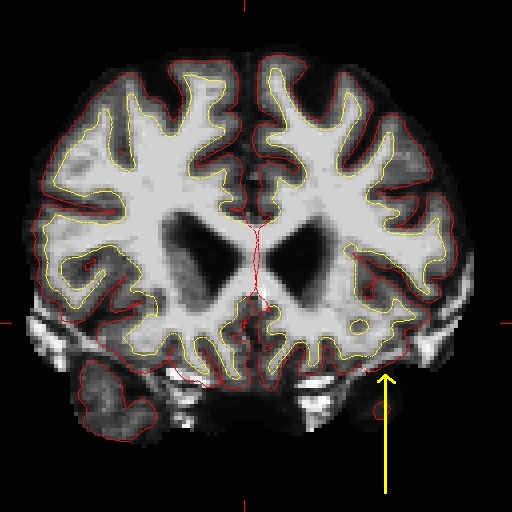
To fix this type of error you can simply edit away the offending voxels from the brainmask.mgz volume. To do this you will need to select the edit voxels tool and set the brush to a size and shape comfortable for you. A circle brush of radius 2 works well for this edit. Then you can simply use the shift key and your mouse button to erase the voxels in the brainmask.mgz, or you can set the brush value to 1 (so that the edits can be detected if you decide to run "recon-all -show-edits") and simple draw over the voxels you want to disappear. At any time you can use the CTRL-Z combination to undo as many actions as you like.
Find a place in the image where the dura is causing errors in the segmentation. Use the right mouse button to delete the voxels. It is not necessary to completely remove the dura to get an adequate pial surface, but it is good to do so until you are more familiar with manual editing. When you are finished removing the bright diagonal line in slice 157, it should look like this:
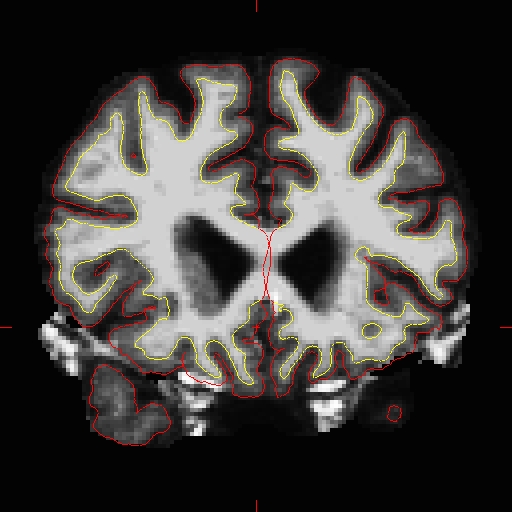
Continue on the other slices until the dura is removed.
When you are done, Click on the wm.mgz, make sure it is highlighted in the menu to the left side of the viewing area and click the save volume button  or save in the file menu.
or save in the file menu.
Regenerating the Surface
When you are finished editing the voxels, you will need to regenerate the surfaces. Since the white matter hasn't been changed, you don't need to resegment the volume. You can regenerate the pial surface with:
recon-all -autorecon-pial -subjid pial_edits_before
Do not run this command if you are conducting the tutorial! This step will take a long time and there is no need to run it for the tutorial purposes.
Edits to correct pial surface extension into cerebellum
The brain.finalsurfs.manedit.mgz volume will allow for the correction of a particular type of problem involving pial surface misplacement whereby parts of the pial surface have extended into the cerebellum in certain areas. It is only in this case where the pial surface extends into cerebellum where the brain.finalsurfs.manedit.mgz volume should be edited. For all other non-cerebellum pial surface problems, brainmask.mgz should be edited. This cerebellum / pial surface problem can be fixed by removing those cerebellum voxels and other surrounding problematic voxels in the brain.finalsurfs.manedit.mgz volume such that upon running recon-all, the pial surface will be pulled in as desired, bordering only gray matter/ CSF boundary, and not jutting into the cerebellum. If only edits to this volume are made on a subject, it is sufficient to re-run recon-all from the point of these edits using the flag -autorecon3-pial. Note that you must create brain.finalsurfs.manedit.mgz by copying it from brain.finalsurfs.mgz:
cd <subjid>/mri cp brain.finalsurfs.mgz brain.finalsurfs.manedit.mgz
You do not need to do this for this tutorial. If you're interested, please refer to the brain.finalsurfs section on the Longitudinal Tutorial here.
Adding control points
Now, take a look at the next subject, cp_before.
freeview -v cp_before/mri/brainmask.mgz cp_before/mri/T1.mgz \ -f cp_before/surf/lh.white:edgecolor=blue cp_before/surf/lh.pial:edgecolor=red \ cp_before/surf/rh.white:edgecolor=blue cp_before/surf/rh.pial:edgecolor=red
Again, this will bring up the brainmask.mgz volume, the T1.mgz volume, and the surfaces for both hemispheres.
In your second terminal window, if not already open, open the surfaces in tksurfer:
tksurfer cp_before lh inflated & tksurfer cp_before rh inflated &
This will open the inflated surfaces for both hemispheres in tksurfer.
The trouble with this subject has occured during the intensity normalization. Check the white surface and be sure it is including all of the white matter as it should. If you find an area that is not included as white matter be sure to check to see that the intensity is at (or very close to) 110.
You may need to open other volumes in order to see or fix the problem.
Using Control Points to Fix Intensity Normalization
Sometimes the intensity normalization step will fail because it cannot determine the proper intensity for white matter. The result is an erroneous white matter segmentation. A control point is a manually selected location in the volume that the user feels sure is inside the white matter boundary, and subsequently should be normalized to an intensity of 110. freeview displays the intensity of any voxel your cursor down below the viewing area, under the "cursor" and "mouse" tables. As you move your mouse around the menu, the value next to T1 and Brainmask should change, it should be 110 when it is hovering over wm.
Manually Selecting Control Points
Subject cp_before is an example of a subject that needs some control points in order to ensure that the voxels are normalized correctly and then included in the wm.mgz volume. This page will walk you through setting control points to fix this type of problem.
freeview -v cp_before/mri/brainmask.mgz cp_before/mri/T1.mgz \ -f cp_before/surf/lh.white:edgecolor=blue cp_before/surf/lh.pial:edgecolor=red \ cp_before/surf/rh.white:edgecolor=blue cp_before/surf/rh.pial:edgecolor=red
Note: If you are using your own subject data, where surfaces are not yet available (ie. the -autorecon2 stage has not run yet), then don't include the surfaces in the freeview command:
freeview -v <my_subj> brainmask.mgz T1.mgz
Scroll through this subject and find the location where the white matter is being excluded from the surface. In subject cp_before, this happens around coronal slice 148.
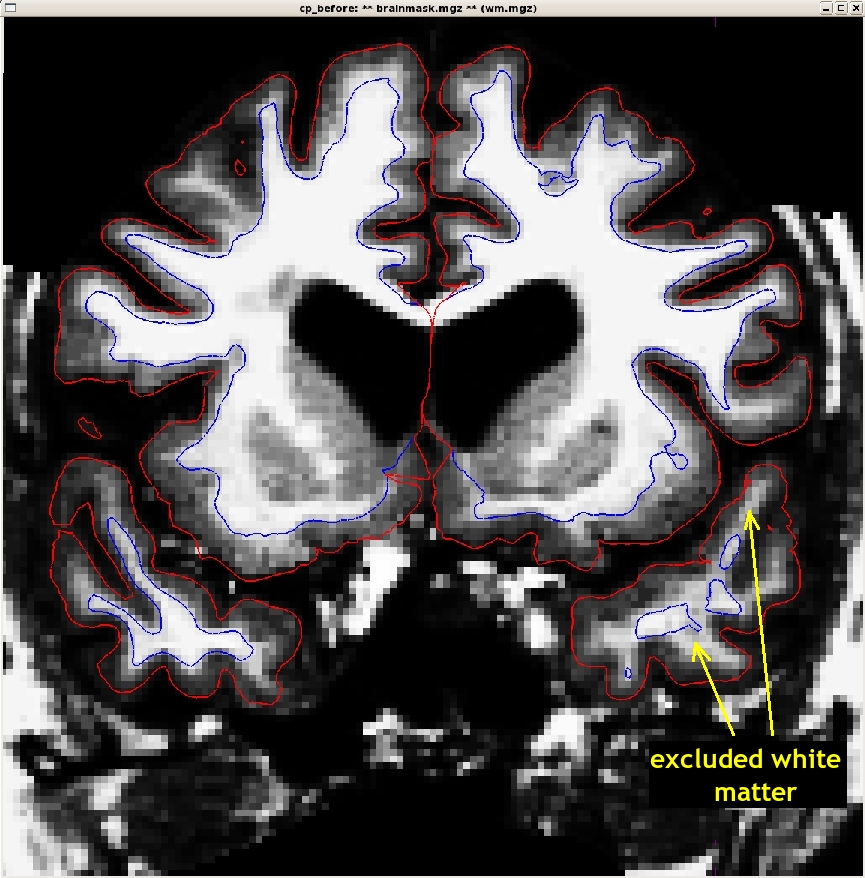
To add control points, you need to click on the "Point Sets" tab to the right of the volume tab. Then click the "New Point Set" tab and name the set "control.dat" make sure the control points option is selected. It should look like this:

Make sure the reference is either the brainmask.mgz or T1.mgz and click "Ok." Use your left mouse button to click where you want to add control points on the volume. Holding down shift while clicking again on a control point will delete it. As you add control points, they will appear as small bright green dots. You can adjust the radius to be smaller if you like. Select a few control points around your trouble areas, space them out throughout the brain and on different slices. You want to pick points in a region where the wm intensity is lower than it should be (that is, having a voxel value less than 110).
General tips for adding control points:
- Control points should only be added in regions that are definitely white matter (i.e., not in the cortex, cerebellum, brainstem, or outside of the skull).
- Control points should also only be added in regions where voxel intensity is not 110. A control point in a region that is already normalized to 110 will be useless.
Control points should NOT be used to try and normalize a brain lesion to 110. Such defects should be fixed with white matter edits
- Control points can help recover thin wm strands that are dark by putting some at the base of the strand.
- Control points are also useful in areas of very bright intensity.
- Start off with a few control points spread out in your trouble area. You may need to add more. With experience you will be able to determine how many are appropriate, given your specific subject.
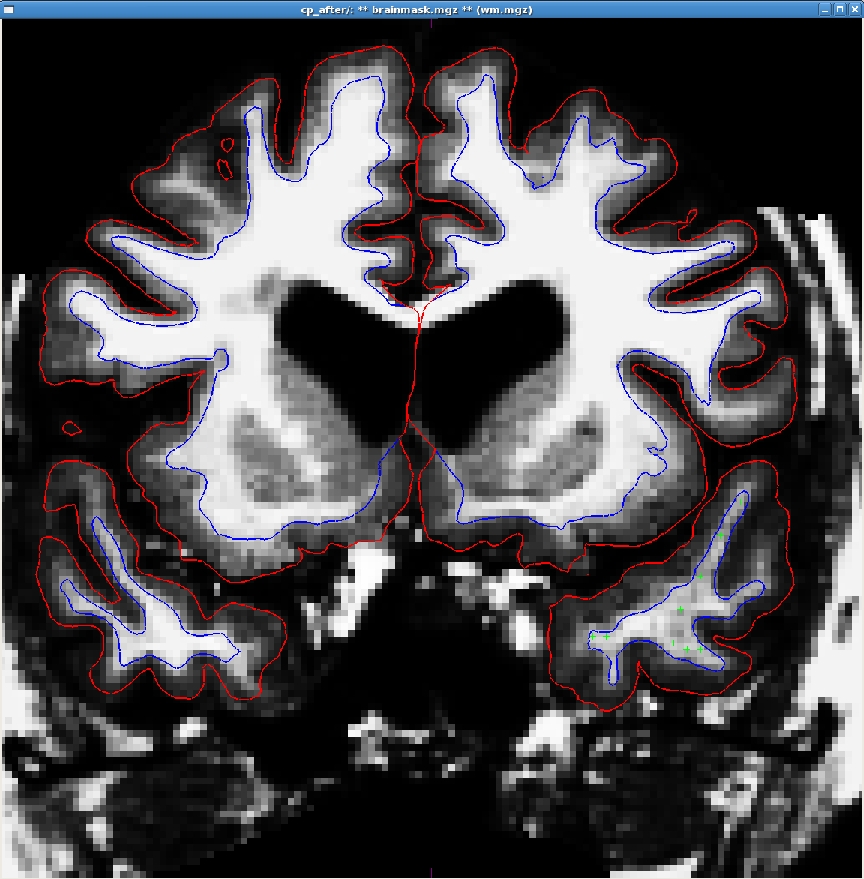
Here is an example of one slice with the control points added. Note that there are other control points spread out through other slices as well.
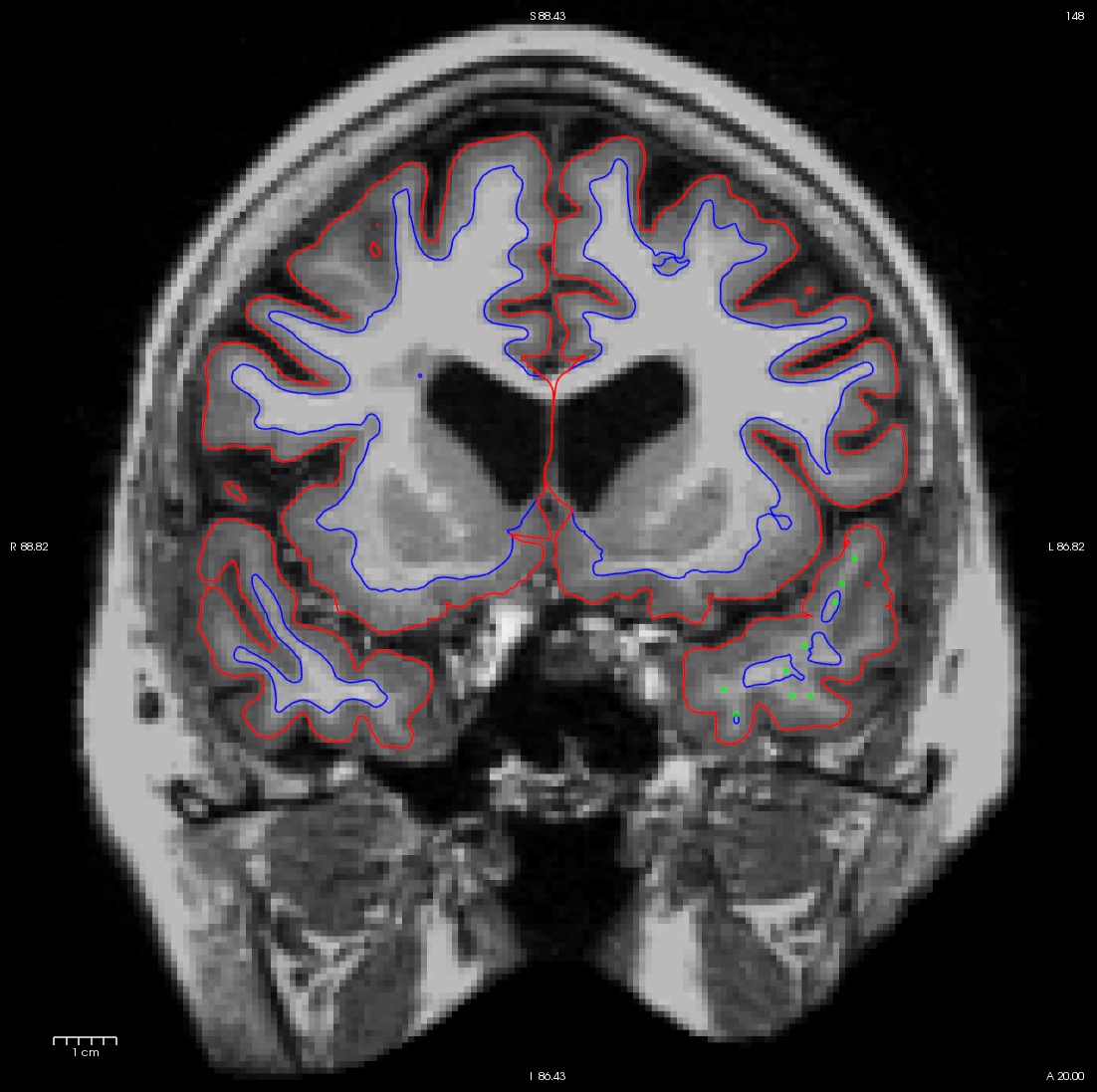
After adding control points, click on the "save point set" button on go to File-> save point set; this will create a file called <subject name>/tmp/control.dat.
Once your control points are saved you can rerun recon-all as follows:
recon-all -autorecon2-cp -autorecon3 -subjid cp_before
Do not run this command if you are conducting the tutorial!
This step will take a long time so there is no need to run it for the purposes of this tutorial.
Using the added control points the subject should look like this after rerunning recon-all: