|
Size: 15645
Comment:
|
Size: 20283
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| [[FsTutorial|Back to Tutorial Top]] <<TableOfContents>> |
## page was renamed from FsTutorial/GroupAnalysis_freeview [[Tutorials|Back to list of all tutorials]] [[FsTutorial|Back to course page]] <<TableOfContents(1)>> |
| Line 9: | Line 13: |
| setenv SUBJECTS_DIR $TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial | export SUBJECTS_DIR=$TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial |
| Line 16: | Line 20: |
| == If You're not at an Organized Course == | == If You're Not at an Organized Course == |
| Line 20: | Line 24: |
| tcsh source your_freesurfer_dir/SetUpFreeSurfer.csh setenv SUBJECTS_DIR $TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial cd $SUBJECTS_DIR/glm }}} Notice the command to open tcsh. If you are already running the tcsh command shell, then the 'tcsh' command is not necessary. If you are not using the tutorial data you should set your {{{SUBJECTS_DIR}}} to the directory in which the recon(s) of the subject(s) you will use for this tutorial are located. |
<source_freesurfer> export TUTORIAL_DATA=<path_to_your_tutorial_data> export SUBJECTS_DIR=$TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial cd $SUBJECTS_DIR/glm }}} If you are not using the tutorial data you should set your {{{SUBJECTS_DIR}}} to the directory in which the recon(s) of the subject(s) you will use for this tutorial are located. |
| Line 29: | Line 33: |
This tutorial is designed to introduce you to the "command-line" group analysis stream in FreeSurfer (as opposed to QDEC which is GUI-driven), including correction for multiple comparisons. While this tutorial shows you how to perform a surface-based thickness study, it is important to realize that most of the concepts learned here apply to any group analysis in FreeSurfer, surface or volume, thickness or fMRI. Here are some useful Group Analysis Links: <<BR>> [[FsTutorial/QdecGroupAnalysis|QDEC Tutorial]]<<BR>> [[FsgdFormat | FSGD Format]]<<BR>> [[FsgdExamples|FSGD Examples]]<<BR>> [[DodsDoss| DODS vs DOSS]]<<BR>> |
This tutorial is designed to introduce you to the "command-line" group analysis stream in FreeSurfer (as opposed to QDEC which is GUI-driven), including correction for multiple comparisons. While this tutorial shows you how to perform a surface-based thickness study, it is important to realize that most of the concepts learned here apply to any group analysis in FreeSurfer, surface or volume, thickness or fMRI. Here are some useful Group Analysis links you might want to refer back to at a later time: <<BR>> [[FsTutorial/QdecGroupAnalysis_freeview|QDEC Tutorial]]<<BR>> [[FsgdFormat|FSGD Format]]<<BR>> [[FsgdExamples|FSGD Examples]]<<BR>> [[DodsDoss|DODS vs DOSS]]<<BR>> |
| Line 44: | Line 38: |
| Line 46: | Line 39: |
The data used for this tutorial is 40 subjects from Randy Buckner's lab. It consists of males and females ages 18 to 93. You can see the demographics [[FsTutorial/GroupAnalysisDemographics|here]]. You will perform an analysis looking for the effect of age on cortical thickness accounting for the effects of gender in the analysis. This is the same data set used in the [[FsTutorial/QdecGroupAnalysis|QDEC Tutorial]], and you will get the same result. |
The data used for this tutorial is 40 subjects from Randy Buckner's lab. It consists of males and females, ages 18 to 93. You can see the demographics [[FsTutorial/GroupAnalysisDemographics|here]]. You will perform an analysis looking for the effect of age on cortical thickness, accounting for the effects of gender in the analysis. This is the same data set used in the [[FsTutorial/QdecGroupAnalysis_freeview|QDEC Tutorial]], and you will get the same result. |
| Line 57: | Line 42: |
| Line 59: | Line 43: |
For this example, we will model the thickness as a straight line. A line has two parameters: intercept (or offset) and a slope. 1. The slope is the change of thickness with age 1. The intercept/offset is interpreted as the thickness at age=0. |
In this tutorial, we will model the thickness as a straight line. A line has two parameters: intercept (or offset) and a slope. For this example: 1. The slope is the change of thickness with age. 1. The intercept/offset is interpreted as the thickness at age=0. |
| Line 66: | Line 50: |
| To account for effects of gender, we will model each sex with its own line, meaning that there will be four linear parameters (also called "betas"): 1. Intercept for Females 1. Intercept for Males 1. Slope for Females 1. Slope for Males In FreeSurfer, this type of design is called [[DodsDoss| DODS]] (for "Different-Offset, Different-Slope"). In FreeSurfer, you can either create your own design matrices, or, if you can specify your design in terms of a FreeSurfer Group Descriptor File [[FsgdFormat| (FSGD)]], FreeSurfer will create them for you. The FSGD file is a simple text file. See [[FsgdFormat|this page]] for the format. The [[FsTutorial/GroupAnalysisDemographics|demographics page]] also has an example FSGD file for this data. |
To account for effects of gender, we will model each sex with its own line, meaning that there will be four linear parameters (also called "betas"): 1. Intercept for Females 1. Intercept for Males 1. Slope for Females 1. Slope for Males In !FreeSurfer, this type of design is called [[DodsDoss|DODS]] (for "Different-Offset, Different-Slope"). You can either create your own design matrices, or, if you specify your design as a !FreeSurfer Group Descriptor File [[FsgdFormat|(FSGD)]], !FreeSurfer will create the design matrices for you. The FSGD file is a simple text file you create. It is not generated by !FreeSurfer. See [[FsgdFormat|this page]] for the format. The [[FsTutorial/GroupAnalysisDemographics|demographics page]] also has an example FSGD file for this data. |
| Line 84: | Line 62: |
| 1. Create an FSGD file for the above design. One (gender_age.fsgd) already exists so that you can continue with the exercises. Information on how to create/view text files can be found [[FsTutorial/CreatingTextfiles|here]]. |
1. Create an FSGD file for the above design. One (gender_age.fsgd) already exists so that you can continue with the exercises. Information on how to create/view text files can be found [[FsTutorial/CreatingTextfiles|here]]. |
| Line 90: | Line 68: |
A contrast is a vector that embodies the hypothesis we want to test. In this case, we wish to test the change in thickness with age after removing the effects of gender. To do this, create a simple text file with the following numbers: |
A contrast is a vector that embodies the hypothesis we want to test. In this case, we wish to test the change in thickness with age, after removing the effects of gender. To do this, create a simple text file with the following numbers: |
| Line 99: | Line 73: |
Notes: 1. There is one value for each parameter (so 4 values) 1. The intercept/offset values are 0 (nuisance) 1. The slope values are 0.5 so as to average the F and M slopes 1. A file called lh-Avg-thickness-age-Cor.mtx already exists = Analysis = == Assemble the Data (mris_preproc) == |
Notes: 1. There is one value for each parameter (so 4 values). 1. The intercept/offset values are 0 (nuisance). 1. The slope values are 0.5 so as to average the F and M slopes. 1. You'll find we created the contrast matrix for you already for this tutorial. It's a file called lh-Avg-thickness-age-Cor.mtx. (You will need to create your own contrast matrix when testing your own hypotheses on your data. FreeSurfer doesn't automatically create this file.) = Assemble the Data (mris_preproc) = |
| Line 111: | Line 82: |
| 1. Resampling each subject's data into a common space, and 1. Concatenating all the subjects' into a single file 1. Spatial smoothing (can be done between 1 and 2) |
1. Resampling each subject's data into a common space. 1. Concatenating all the subjects' into a single file. 1. Spatial smoothing (can be done between 1 and 2). |
| Line 116: | Line 89: |
| === Previously Cached (qcached) Data === When you have run recon-all with the -qcache option, recon-all will resample data onto the average subject (fsaverage) and smooth it at various FWHM (full-width/half-max), usually 0, 5, 10, 10, 20, and 25mm. This can speed later processing. The data for this tutorial have been cached, so run: ---- |
== Previously Cached (qcached) Data == When you run recon-all with the -qcache option, recon-all will resample data onto the average subject (called fsaverage) and smooth it at various FWHM (full-width/half-max) values, usually 0, 5, 10, 15, 20, and 25mm. This can speed later processing. The data for this tutorial was ran with the -qcache option, so the FWHM are already cached. Run this command to assemble your previously cached data: |
| Line 127: | Line 95: |
| --target fsaverage --hemi lh \ | --target fsaverage \ --hemi lh \ |
| Line 129: | Line 98: |
| Line 132: | Line 100: |
Notes: 1. Only takes a few seconds because the data have been cached 1. The FSGD file lists all the subjects, helps keep order. 1. The independent variable is the thickness smoothed to 10mm FWHM. 1. The data are for the left hemisphere 1. The output is lh.gender_age.thickness.10.mgh (which already exists) |
'''NOTE''': The backslash allows you to copy and paste multiple lines of code as one command. We use this throughout the tutorials to display the commands in a more easy-to-read manner, while still allowing you to copy and paste. Whenever you are typing in your own commands, instead of copying and pasting a command written out across multiple lines, a backslash is not necessary. ---- Notes: 1. Only takes a few seconds because the data have been cached. 1. The FSGD file lists all the subjects, helps keep order. 1. The independent variable is the thickness smoothed to 10mm FWHM. 1. The target is the average subject, fsaverage. This subject is included with the !FreeSurfer distribution. 1. The data are for the left hemisphere. 1. Since the data is already cached, this {{{mris_preproc}}} command just creates a stack of all the smoothed thickness data from each subject into one file as output. 1. The output of this command is given by the --out flag. Here we call it '''lh.gender_age.thickness.10.mgh'''. |
| Line 141: | Line 114: |
| 1. Run mri_info on lh.gender_age.thickness.10.mgh to find its dimensions,ie, ---- |
1. Run mri_info on lh.gender_age.thickness.10.mgh to find its dimensions: |
| Line 147: | Line 120: |
| ---- === Uncached Data === In the case that you have not cached the data, you can perform the same operations manually using the two commands below. There is no need to do this for this tutorial. OPTIONAL: THIS WILL TAKE ABOUT 10 MINUTES |
== Uncached Data == In the case that you have not cached the data, you can perform the same operations manually using the two commands below. These two commands give the equivalent results as running recon-all with -qcache. However, the commands below only create output for unsmoothed data and data smoothed to 10mm FWHM. qcache, on the other hand, smooths the data for a range of different FWHM vaules. '''THESE NEXT TWO COMMANDS ARE OPTIONAL FOR THE TUTORIAL, AND WILL TAKE ABOUT 10 MINUTES TO RUN. ''' ''' The data has already been pre-processed for you.''' |
| Line 157: | Line 129: |
| --target fsaverage --hemi lh \ | --target fsaverage \ --hemi lh \ |
| Line 161: | Line 134: |
Notes: 1. This resamples each subjects data to fsaverage 1. Output is lh.gender_age.thickness.00.mgh, which is unsmoothed OPTIONAL: THIS WILL TAKE ABOUT 5 MINUTES |
Notes: 1. This resamples each subject's left hemisphere data to fsaverage. 1. Output is lh.gender_age.thickness.00.mgh, which is unsmoothed. ##'''OPTIONAL: THIS WILL TAKE ABOUT 5 MINUTES''' |
| Line 177: | Line 148: |
1. This smooths by 10mm FWHM 1. "--cortex" means only smooth areas in cortex (exclude medial wall). This is automatically done with qcache. You can also specify other labels. 1. Output is lh.gender_age.thickness.10B.mgh. == GLM Analysis (mri_glmfit) == ---- |
1. This smooths each subject's resampled data by 10mm FWHM. 1. "--cortex" means only smooth areas in cortex (exclude medial wall). This is automatically done with qcache. You can also specify other labels. 1. Output is lh.gender_age.thickness.10B.mgh. (We have named the output file ...10B.mgh because the precreated file from running qcache ...10.mgh already exists.) = GLM Analysis (mri_glmfit) = |
| Line 195: | Line 161: |
}}} ---- Notes: 1. Input is lh.gender_age.thickness.10.mgh 1. Same FSGD used as with mris_preproc. Maintains subject order! 1. DODS is specified (it is the default) 1. Only one contrast is used (lh-Avg-thickness-age-Cor.mtx) but you can specify multiple contrasts. 1. "--cortex" specifies that the analysis only be done in cortex (ie, medial wall is zeroed out). Other labels can be used. 1. The output directory is lh.gender_age.glmdir 1. Should only take about 1min to run. |
}}} Notes: 1. Input is lh.gender_age.thickness.10.mgh. 1. Same FSGD used as with mris_preproc. Maintains subject order! 1. DODS is specified (it is the default). 1. Only one contrast is used (lh-Avg-thickness-age-Cor.mtx), but you can specify multiple contrasts. 1. "--cortex" specifies that the analysis only be done in cortex (ie, medial wall is zeroed out). Other labels can be used. 1. The output directory is lh.gender_age.glmdir. 1. Should only take about 1min to run. |
| Line 211: | Line 174: |
| When this command is finished you will have an lh.gender_age.glmdir. There will be a number of output files in this directory, as well as two other directories. If you did an ''ls'' in your glmdir there will be: {{{ |
When this command is finished the '''glm''' directory will contain a sub-directory called '''lh.gender_age.glmdir'''. There will be a number of output files in this directory, as well as other subdirectories. Run: {{{ ls lh.gender_age.glmdir }}} and you will see the following files (descriptions are also included below, but don't show up in your terminal screen): {{{ beta.mgh -- all parameter estimates (surface overlay) dof.dat -- degrees of freedom (text) fwhm.dat -- average FWHM of residual (text) lh-Avg-thickness-age-Cor -- contrast subdirectory mask.mgh -- binary mask (surface overlay) mri_glmfit.log -- log file (text, send this with bug reports) rstd.mgh -- residual standard deviation (surface overlay) rvar.mgh -- residual variance (surface overlay) sar1.mgh -- residual spatial AR1 (surface overlay) surface -- the subject and hemisphere used for this analysis (text) Xg.dat -- design matrix (text) X.mat -- design matrix (MATLAB format) |
| Line 218: | Line 195: |
| Xg.dat -- design matrix (text) mask.mgh -- binary mask (surface overlay) beta.mgh -- all parameter estimates (surface overlay) rstd.mgh -- residual standard deviation (surface overlay) sar1.mgh -- residual spatial AR1 (surface overlay) fwhm.dat -- average FWHM of residual (text) dof.dat -- degrees of freedom (text) lh-Avg-thickness-age-Cor -- contrast subdirectory mri_glmfit.log -- log file (text, send this with bug reports) }}} |
}}} '''NOTE:''' You may also see some temporary directories (with names starting with tmp) or log files, which you can ignore. |
| Line 230: | Line 199: |
| 1. What was the DOF for this experiment? 1. What was the FWHM? 1. How many frames does beta have? Why? There will be a subdirectory for each contrast that you specify. The name of the directory will be that of the contrast matrix file (without the .mtx extension). The '''lh-Avg-thickness-age-Cor''' directory will have the following files: |
1. What was the DOF for this experiment? 1. What was the FWHM? 1. How many frames does beta have? Why? (hint: run mri_info on lh.gender_age.glmdir/beta.mgh) There will be a subdirectory for each contrast that you specify. The name of the directory will be that of the contrast matrix file (without the .mtx extension). For example, to inspect the '''lh-Avg-thickness-age-Cor''' directory, run {{{ ls lh.gender_age.glmdir/lh-Avg-thickness-age-Cor }}} and you will see the following files: |
| Line 241: | Line 213: |
| cnr.mgh -- contrast-to-noise ratio (surface overlay) efficiency.dat -- statistical efficiency for the contrast (text) F.mgh -- F ratio of contrast (surface overlay) |
|
| Line 242: | Line 217: |
| F.mgh -- F ratio of contrast (surface overlay) | gammavar.mgh --contrast variance (surface overlay) maxvox.dat -- voxel with the maximum statistic (text) pcc.mgh -- partial (pearson) correlation coefficient (surface overlay) |
| Line 244: | Line 221: |
| }}} View the uncorrected significance map with tksurfer: ---- {{{ tksurfer fsaverage lh inflated \ -annot aparc.annot -fthresh 2 \ -overlay lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/sig.mgh }}} ---- You should see: {{attachment:glm_1.jpg}} {{attachment:glm_2.jpg}} Notes: 1. The threshold is set to 2, meaning vertices with p<.01, uncorrected, will have color. 1. Blue means a negative correlation (thickness decreases with age), red is positive correlation. 1. Click on a point in the Precentral Gyrus. What is it's value? What does it mean? Viewing the medial surface, change the overlay threshold to something very, very low (say, .01): {{{ View->Configure->Overlay, set "Min" .01 }}} You should see: {{attachment:glm_3.jpg}} Notes: 1. Almost all of cortex now has color 1. The non-cortical areas are still blank (0) because they were excluded with --cortex in mri_glmfit above. == Clusterwise Correction for Multiple Comparisons == |
z.mgh -- z-stat that corresponds to the significance (surface overlay) }}} View the uncorrected significance map with freeview. First, make sure you are in the correct directory: {{{ cd $SUBJECTS_DIR/glm }}} Then, run this command to visualize the data: {{{ freeview -f $SUBJECTS_DIR/fsaverage/surf/lh.inflated:annot=aparc.annot:annot_outline=1:overlay=lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/sig.mgh:overlay_threshold=4,5 -viewport 3d }}} You should see something similar to this: {{attachment:glm1_fv5.3.jpg||height="370"}} {{attachment:glm2_fv5.3.jpg||height="370"}} Notes: 1. The threshold is set to 4, meaning vertices with p<.0001, uncorrected, will have color. 1. Blue means a negative correlation (thickness decreases with age), red is positive correlation. 1. Click on a point in the Precentral Gyrus. What is its value? What does it mean? Viewing the medial surface, change the overlay threshold to something very, very low (say, .01), by clicking '''Configure''' (under the Overlay dropdown menu that says sig.mgh) and set the '''Min''' value to 0.01. Click Apply, or check the '''Automatically apply changes''' checkbox. You should see something like this: {{attachment:glm3_fv5.3.jpg||height="370"}} Notes: 1. Almost all of the cortex now has color. 1. The non-cortical areas are still blank (0) because they were excluded with --cortex in mri_glmfit above. All the surface overlays created by mri_glmfit, not just the significance map, can also be inspected in freeview. Simply replace the path to the sig.mgh file in the command above to the surface overlay that you would like to see. For example, this will display the F ratio instead of the significance map: {{{ freeview -f $SUBJECTS_DIR/fsaverage/surf/lh.inflated:annot=aparc.annot:annot_outline=1:overlay=lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/F.mgh:overlay_threshold=20,50 -viewport 3d }}} Text output and log files can be opened in any text editor. = Clusterwise Correction for Multiple Comparisons = |
| Line 288: | Line 264: |
| To perform a cluster-wise correction for multiple comparisons, we will run a simulation. The simulation is a way to get a measure of the distribution of the maximum cluster size under the null hypothesis. This is done by iterating over the following steps: 1. Synthesize a z map 1. Smooth z map (using residual FWHM) 1. Threshold z map (level and sign) 1. Find clusters in thresholded map 1. Record area of maximum cluster 1. Repeat over desired number of iterations (usually > 5000) In FreeSurfer, this information is stored in a simple text file called a CSD (Cluster Simulation Data). Once we have the distribution of the maximum cluster size, we correct for multiple comparisons by: 1. Going back to the original data 1. Thresholding using same level and sign 1. Finding clusters in thresholded map 1. For each cluster, p = probability of seeing a maximum cluster that size or larger during simulation. === Run the simulation === All these steps are performed with the mri_glmfit-sim: ---- |
==== Remember to run the following commands in every new terminal window (aka shell) you open throughout this tutorial (see top of page for detailed instructions): ==== If You're at an Organized Course: {{{ export SUBJECTS_DIR=$TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial cd $SUBJECTS_DIR/glm }}} If You're Not at an Organized Course: {{{ <source_freesurfer> export TUTORIAL_DATA=<path_to_your_tutorial_data> export SUBJECTS_DIR=$TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial cd $SUBJECTS_DIR/glm }}} -------- To perform a cluster-wise correction for multiple comparisons, we will run a simulation. The simulation is a way to get a measure of the distribution of the maximum cluster size under the null hypothesis. This is done by iterating over the following steps: 1. Synthesize a z map. 1. Smooth z map (using residual FWHM). 1. Threshold z map (level and sign). 1. Find clusters in thresholded map. 1. Record area of maximum cluster. 1. Repeat over desired number of iterations (usually > 5,000). In !FreeSurfer, this information is stored in a simple text file called a CSD (Cluster Simulation Data) file. Once we have the distribution of the maximum cluster size, we correct for multiple comparisons by: 1. Going back to the original data. 1. Thresholding using same level and sign. 1. Finding clusters in thresholded map. 1. For each cluster, p = probability of seeing a maximum cluster that size or larger during simulation. == Run the simulation == All these steps are performed with the command mri_glmfit-sim: |
| Line 318: | Line 304: |
| --sim mc-z 5 4 mc-z.negative \ --sim-sign neg --cwpvalthresh 0.4\ --overwrite }}} ---- Notes: 1. Specify the same GLM directory (--glmdir) 1. The simulation type is Z Monte Carlo (mc-z) 1. Specify the sign (neg for negative, pos, or abs) 1. Vertex-wise threshold of 4 (p < .0001) 1. Only 5 iterations (usually > 5000) 1. CSD files will have mc-z.negative base 1. "--cwpvalthresh 0.4" Keep clusters that have cluster-wise p-values < 0.4 (usually you will want something like .05). To see all clusters, set to .999 1. --overwrite deletes previous CSD files This has been run this data with 1000 iterations (CSD base of mc-z.neg). This can often take many hours. === View CSD File === The CSD files are stored in lh.gender_age.glmdir/csd. Look at one of them: ---- {{{ less lh.gender_age.glmdir/csd/mc-z.neg4.j001-lh-Avg-thickness-age-Cor.csd }}} ---- Or click [[FsTutorial/GroupAnalysisCsd|here]]. ''To end the {{{less}}} command, type 'q' for quit.'' Notes: 1. Rows that begin with '#' are headers or comments. 1. Each data row is an iteration. 1. The 3rd column is the maximum cluster size for that iteration. 1. The CSD file itself is not particularly important, so don't worry too much about it. === View the Corrected Results === In the contrast subdirectory, you will see several new files, all beginning with mc-z: {{{ mc-z.neg4.sig.cluster.summary - summary of clusters (text) mc-z.neg4.sig.cluster.mgh - cluster-wise corrected map (overlay) mc-z.neg4.sig.ocn.annot - annotation of clusters }}} First, look at the cluster summary (or click [[FsTutorial/GroupAnalysisClusterSummary|here]]): ---- {{{ less lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/mc-z.neg4.sig.cluster.summary }}} ---- Notes: 1. This is a list of all the clusters that were found (38 of them) 1. The CWP column is the cluster-wise probability (the number you are interested in). It is a simple p (ie, NOT -log10(p)). 1. Note that ALL clusters are found, regardless of their significance. 1. Cluster number 1 has a CWP of p=.001. 1. Cluster number 21 has a CWP of p=.077. Load the cluster annotation in tksurfer ---- {{{ tksurfer fsaverage lh inflated \ -annot lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/mc-z.neg4.sig.ocn.annot \ -fthresh 2 -curv -gray }}} ---- You should see: {{attachment:lateral_clusters.jpg}} {{attachment:medial_clusters.jpg}} Notes: 1. These are all clusters, regardless of significance. 1. When you click on a cluster, the label will tell you the cluster number (eg, cluster-021). Change the label mode to "Outline" by hitting the outline button {{attachment:tksurfer.outline.gif}}. Now load the cluster p-value overlay with {{{ File->LoadOverlay Click "Browse..." Next to where it says "Selection", paste (middle click with a 3-button mouse) the following: lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/mc-z.neg4.sig.cluster.mgh Hit OK Hit OK }}} You should see: {{attachment:lateral.jpg}} {{attachment:medial.jpg}} Note: |
--cache 4 neg \ --cwp 0.05\ --2spaces }}} Notes: 1. Specify the same GLM directory (--glmdir). 1. Use precomputed Z Monte Carlo simulation (--cache). 1. Vertex-wise/cluster-forming threshold of 4 (p < .0001). 1. Specify the sign ("neg" for negative, "pos" for positive, or "abs" for absolute/unsigned). 1. --cwp 0.05 : Keep clusters that have cluster-wise p-values < 0.05. To see all clusters, set to .999. 1. --2spaces : adjust p-values for two hemispheres (this assumes you will eventually look at the right hemisphere too). 1. You can also generate your own precomputed data, but you only need to do this if you have a different average subject or you are not looking over all of the cortex. 1. You can also use a permutation simulation instead of Z Monte Carlo. Run mri_glmfit-sim --help to get more information. == View the Corrected Results == In the contrast subdirectory, you will see several new files by running: {{{ ls lh.gender_age.glmdir/lh-Avg-thickness-age-Cor }}} You will see the following new files: {{{ cache.th40.neg.sig.pdf.dat -- probability distribution function of clusterwise correction cache.th40.neg.sig.cluster.mgh -- cluster-wise corrected map (overlay) cache.th40.neg.sig.cluster.summary -- summary of clusters (text) cache.th40.neg.sig.masked.mgh -- uncorrected sig values masked by the clusters that survive correction cache.th40.neg.sig.ocn.annot -- output cluster number (annotation of clusters) cache.th40.neg.sig.ocn.mgh -- output cluster number (segmentation showing where each numbered cluster is) cache.th40.neg.sig.voxel.mgh -- voxel-wise map corrected for multiple comparisons at a voxel (rather than cluster) level cache.th40.neg.sig.y.ocn.dat -- the average value of each subject in each cluster }}} First, look at the cluster summary (or click [[FsTutorial/GroupAnalysisClusterSummary|here]]): {{{ less lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/cache.th40.neg.sig.cluster.summary }}} You can hit the 'Page Up' and 'Page Down' buttons or the 'Up' and 'Down' arrow keys to see the rest of the file. '''(To exit the less command, hit the 'q' button.)''' Notes: 1. This is a list of all the clusters that were found (20 of them). 1. The CWP column is the cluster-wise probability (the number you are interested in). It is a simple p (ie, NOT -log10(p)). 1. For example, cluster number 1 has a CWP of p=.0002. Load the cluster annotation in freeview: {{{ freeview -f $SUBJECTS_DIR/fsaverage/surf/lh.inflated:overlay=lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/cache.th40.neg.sig.cluster.mgh:overlay_threshold=2,5:annot=lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/cache.th40.neg.sig.ocn.annot -viewport 3d }}} You should see clusters similar in shape to those pictured in the snapshots below. The color values associated with each cluster are arbitrary and may be different: {{attachment:multiple_comparisons_lateral.jpg||height="370"}} {{attachment:multiple_comparisons_medial.jpg||height="370"}} Notes: 1. These are all clusters, regardless of significance. 1. When you click on a cluster, the label will tell you the cluster number (eg, cluster-016). |
| Line 423: | Line 366: |
| 1. Find and click on cluster 1. It is blue and has a value of -3 since this is log10(.001). The -3 is because the correlation is negative. 1. Find and click on cluster 21. Its value is -1.11 because this is log10(.077). Note that it is transparent because its significance is worse than the threshold we set (-fthresh 2, p < .01). 1. All vertices within a cluster are the same value (the p-value of the cluster). 1. You can changed the cluster-wise threshold by clicking on View->Configure->Overlay, and setting the "Min" to your desired level. As you do this, some clusters will change transparency. |
1. Find and click on cluster 1 (the largest cluster). It has a value of -3.69899 since this is log10(.0002). The -3.69899 is because the correlation is negative. 1. Find and click on cluster 17 (on the medial side of the brain). Its value is -1.4023 because this is log10(0.03960). Note that if you turn off the annotation, the cluster 17 is not visible because its significance is worse than the threshold we set (-fthresh 2, p < .01). 1. All vertices within a cluster are the same value (the p-value of the cluster). 1. You can change the cluster-wise threshold by first clicking on "Show outline only" underneath the Annotation drop down menu. Then click on '''Configure '''(underneath "Overlay"), and set the '''Min''' value to your desired level. Alternatively, you can drag the red flag to adjust the cluster-wise threshold. As you do this, clusters will appear or disappear from the surface. (If your cursor is in the Min text box, the red flag won't move. Click on another text box to be able to move the flag.) [[https://surfer.nmr.mgh.harvard.edu/fswiki/Tutorials|Back to list of all tutorials]] [[FsTutorial|Back to course page]] |
Contents
Preparations
If You're at an Organized Course
If you are taking one of the formally organized courses, everything has been set up for you on the provided laptop. The only thing you will need to do is run the following commands in every new terminal window (aka shell) you open throughout this tutorial. Copy and paste the commands below to get started:
export SUBJECTS_DIR=$TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial cd $SUBJECTS_DIR/glm
To copy: Highlight the command in the box above, right click and select copy (or use keyboard shortcut Ctrl+c), then use the middle button of your mouse to click inside the terminal window (this will paste the command). Press enter to run the command.
These two commands set the SUBJECTS_DIR variable to the directory where the data is stored and then navigates into this directory. You can now skip ahead to the tutorial (below the gray line).
If You're Not at an Organized Course
If you are NOT taking one of the formally organized courses, then to follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names. These are the commands that you need to run before getting started:
<source_freesurfer> export TUTORIAL_DATA=<path_to_your_tutorial_data> export SUBJECTS_DIR=$TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial cd $SUBJECTS_DIR/glm
If you are not using the tutorial data you should set your SUBJECTS_DIR to the directory in which the recon(s) of the subject(s) you will use for this tutorial are located.
Introduction
This tutorial is designed to introduce you to the "command-line" group analysis stream in FreeSurfer (as opposed to QDEC which is GUI-driven), including correction for multiple comparisons. While this tutorial shows you how to perform a surface-based thickness study, it is important to realize that most of the concepts learned here apply to any group analysis in FreeSurfer, surface or volume, thickness or fMRI.
Here are some useful Group Analysis links you might want to refer back to at a later time:
QDEC Tutorial
FSGD Format
FSGD Examples
DODS vs DOSS
This Data Set
The data used for this tutorial is 40 subjects from Randy Buckner's lab. It consists of males and females, ages 18 to 93. You can see the demographics here. You will perform an analysis looking for the effect of age on cortical thickness, accounting for the effects of gender in the analysis. This is the same data set used in the QDEC Tutorial, and you will get the same result.
General Linear Model (GLM) DODS Setup
Design Matrix/FSGD File
In this tutorial, we will model the thickness as a straight line. A line has two parameters: intercept (or offset) and a slope. For this example:
- The slope is the change of thickness with age.
- The intercept/offset is interpreted as the thickness at age=0.
Parameter estimates are also called "regression coefficients".
To account for effects of gender, we will model each sex with its own line, meaning that there will be four linear parameters (also called "betas"):
- Intercept for Females
- Intercept for Males
- Slope for Females
- Slope for Males
In FreeSurfer, this type of design is called DODS (for "Different-Offset, Different-Slope").
You can either create your own design matrices, or, if you specify your design as a FreeSurfer Group Descriptor File (FSGD), FreeSurfer will create the design matrices for you. The FSGD file is a simple text file you create. It is not generated by FreeSurfer. See this page for the format. The demographics page also has an example FSGD file for this data.
Things to do:
- Create an FSGD file for the above design. One (gender_age.fsgd) already exists so that you can continue with the exercises.
Information on how to create/view text files can be found here.
Contrasts
A contrast is a vector that embodies the hypothesis we want to test. In this case, we wish to test the change in thickness with age, after removing the effects of gender. To do this, create a simple text file with the following numbers:
0 0 0.5 0.5
Notes:
- There is one value for each parameter (so 4 values).
- The intercept/offset values are 0 (nuisance).
- The slope values are 0.5 so as to average the F and M slopes.
You'll find we created the contrast matrix for you already for this tutorial. It's a file called lh-Avg-thickness-age-Cor.mtx. (You will need to create your own contrast matrix when testing your own hypotheses on your data. FreeSurfer doesn't automatically create this file.)
Assemble the Data (mris_preproc)
Assembling the data simply means:
- Resampling each subject's data into a common space.
- Concatenating all the subjects' into a single file.
- Spatial smoothing (can be done between 1 and 2).
This can be done in two equivalent ways:
Previously Cached (qcached) Data
When you run recon-all with the -qcache option, recon-all will resample data onto the average subject (called fsaverage) and smooth it at various FWHM (full-width/half-max) values, usually 0, 5, 10, 15, 20, and 25mm. This can speed later processing. The data for this tutorial was ran with the -qcache option, so the FWHM are already cached. Run this command to assemble your previously cached data:
mris_preproc --fsgd gender_age.fsgd \ --cache-in thickness.fwhm10.fsaverage \ --target fsaverage \ --hemi lh \ --out lh.gender_age.thickness.10.mgh
NOTE: The backslash allows you to copy and paste multiple lines of code as one command. We use this throughout the tutorials to display the commands in a more easy-to-read manner, while still allowing you to copy and paste. Whenever you are typing in your own commands, instead of copying and pasting a command written out across multiple lines, a backslash is not necessary.
Notes:
- Only takes a few seconds because the data have been cached.
- The FSGD file lists all the subjects, helps keep order.
- The independent variable is the thickness smoothed to 10mm FWHM.
The target is the average subject, fsaverage. This subject is included with the FreeSurfer distribution.
- The data are for the left hemisphere.
Since the data is already cached, this mris_preproc command just creates a stack of all the smoothed thickness data from each subject into one file as output.
The output of this command is given by the --out flag. Here we call it lh.gender_age.thickness.10.mgh.
Things to do:
- Run mri_info on lh.gender_age.thickness.10.mgh to find its dimensions:
mri_info lh.gender_age.thickness.10.mgh
Uncached Data
In the case that you have not cached the data, you can perform the same operations manually using the two commands below. These two commands give the equivalent results as running recon-all with -qcache. However, the commands below only create output for unsmoothed data and data smoothed to 10mm FWHM. qcache, on the other hand, smooths the data for a range of different FWHM vaules.
THESE NEXT TWO COMMANDS ARE OPTIONAL FOR THE TUTORIAL, AND WILL TAKE ABOUT 10 MINUTES TO RUN.
The data has already been pre-processed for you.
mris_preproc --fsgd gender_age.fsgd \ --target fsaverage \ --hemi lh \ --meas thickness \ --out lh.gender_age.thickness.00.mgh
Notes:
- This resamples each subject's left hemisphere data to fsaverage.
- Output is lh.gender_age.thickness.00.mgh, which is unsmoothed.
mri_surf2surf --hemi lh \ --s fsaverage \ --sval lh.gender_age.thickness.00.mgh \ --fwhm 10 \ --cortex \ --tval lh.gender_age.thickness.10B.mgh
- This smooths each subject's resampled data by 10mm FWHM.
- "--cortex" means only smooth areas in cortex (exclude medial wall). This is automatically done with qcache. You can also specify other labels.
- Output is lh.gender_age.thickness.10B.mgh. (We have named the output file ...10B.mgh because the precreated file from running qcache ...10.mgh already exists.)
GLM Analysis (mri_glmfit)
mri_glmfit \ --y lh.gender_age.thickness.10.mgh \ --fsgd gender_age.fsgd dods\ --C lh-Avg-thickness-age-Cor.mtx \ --surf fsaverage lh \ --cortex \ --glmdir lh.gender_age.glmdir
Notes:
- Input is lh.gender_age.thickness.10.mgh.
- Same FSGD used as with mris_preproc. Maintains subject order!
- DODS is specified (it is the default).
- Only one contrast is used (lh-Avg-thickness-age-Cor.mtx), but you can specify multiple contrasts.
- "--cortex" specifies that the analysis only be done in cortex (ie, medial wall is zeroed out). Other labels can be used.
- The output directory is lh.gender_age.glmdir.
- Should only take about 1min to run.
Things to do:
When this command is finished the glm directory will contain a sub-directory called lh.gender_age.glmdir. There will be a number of output files in this directory, as well as other subdirectories. Run:
ls lh.gender_age.glmdir
and you will see the following files (descriptions are also included below, but don't show up in your terminal screen):
beta.mgh -- all parameter estimates (surface overlay) dof.dat -- degrees of freedom (text) fwhm.dat -- average FWHM of residual (text) lh-Avg-thickness-age-Cor -- contrast subdirectory mask.mgh -- binary mask (surface overlay) mri_glmfit.log -- log file (text, send this with bug reports) rstd.mgh -- residual standard deviation (surface overlay) rvar.mgh -- residual variance (surface overlay) sar1.mgh -- residual spatial AR1 (surface overlay) surface -- the subject and hemisphere used for this analysis (text) Xg.dat -- design matrix (text) X.mat -- design matrix (MATLAB format) y.fsgd -- copy of input FSGD file (text)
NOTE: You may also see some temporary directories (with names starting with tmp) or log files, which you can ignore.
Study Questions:
- What was the DOF for this experiment?
- What was the FWHM?
- How many frames does beta have? Why? (hint: run mri_info on lh.gender_age.glmdir/beta.mgh)
There will be a subdirectory for each contrast that you specify. The name of the directory will be that of the contrast matrix file (without the .mtx extension). For example, to inspect the lh-Avg-thickness-age-Cor directory, run
ls lh.gender_age.glmdir/lh-Avg-thickness-age-Cor
and you will see the following files:
C.dat -- original contrast matrix (text) cnr.mgh -- contrast-to-noise ratio (surface overlay) efficiency.dat -- statistical efficiency for the contrast (text) F.mgh -- F ratio of contrast (surface overlay) gamma.mgh -- contrast effect size (surface overlay) gammavar.mgh --contrast variance (surface overlay) maxvox.dat -- voxel with the maximum statistic (text) pcc.mgh -- partial (pearson) correlation coefficient (surface overlay) sig.mgh -- significance, -log10(pvalue), uncorrected (surface overlay) z.mgh -- z-stat that corresponds to the significance (surface overlay)
View the uncorrected significance map with freeview. First, make sure you are in the correct directory:
cd $SUBJECTS_DIR/glm
Then, run this command to visualize the data:
freeview -f $SUBJECTS_DIR/fsaverage/surf/lh.inflated:annot=aparc.annot:annot_outline=1:overlay=lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/sig.mgh:overlay_threshold=4,5 -viewport 3d
You should see something similar to this:
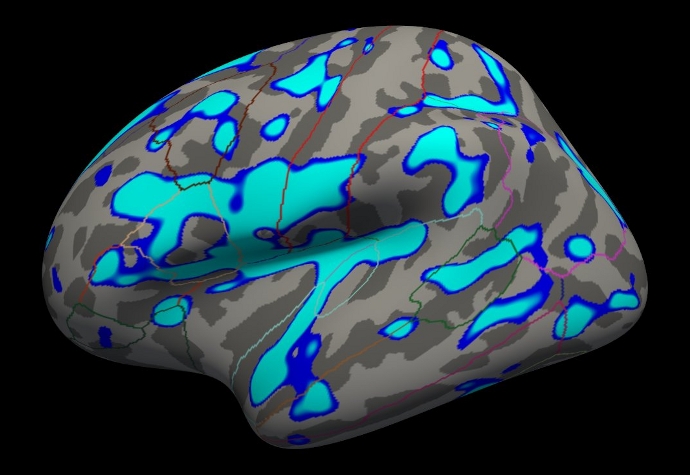
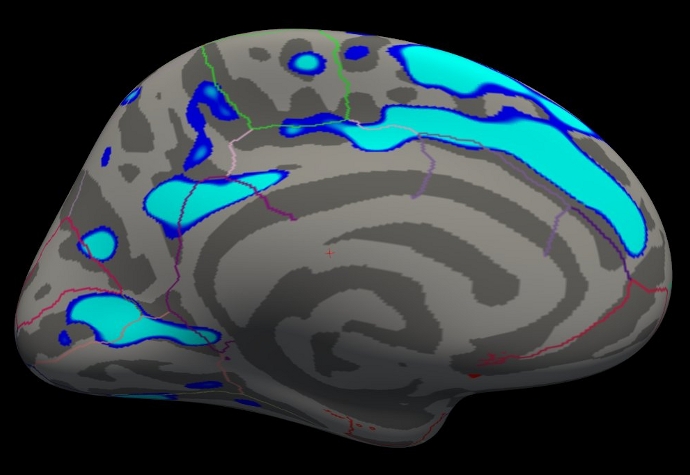
Notes:
The threshold is set to 4, meaning vertices with p<.0001, uncorrected, will have color.
- Blue means a negative correlation (thickness decreases with age), red is positive correlation.
- Click on a point in the Precentral Gyrus. What is its value? What does it mean?
Viewing the medial surface, change the overlay threshold to something very, very low (say, .01), by clicking Configure (under the Overlay dropdown menu that says sig.mgh) and set the Min value to 0.01. Click Apply, or check the Automatically apply changes checkbox.
You should see something like this:
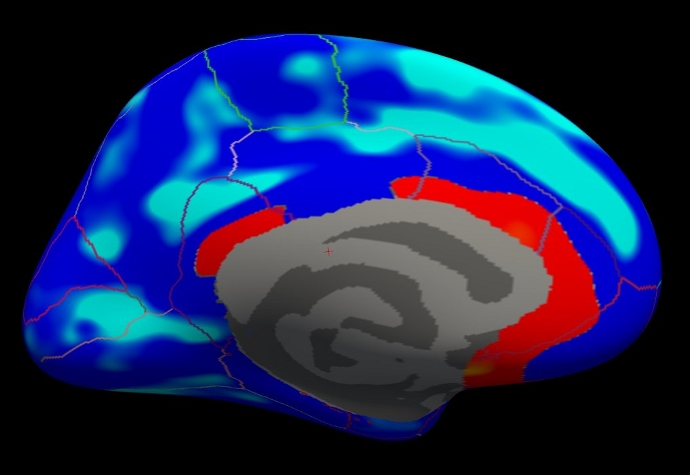
Notes:
- Almost all of the cortex now has color.
- The non-cortical areas are still blank (0) because they were excluded with --cortex in mri_glmfit above.
All the surface overlays created by mri_glmfit, not just the significance map, can also be inspected in freeview. Simply replace the path to the sig.mgh file in the command above to the surface overlay that you would like to see. For example, this will display the F ratio instead of the significance map:
freeview -f $SUBJECTS_DIR/fsaverage/surf/lh.inflated:annot=aparc.annot:annot_outline=1:overlay=lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/F.mgh:overlay_threshold=20,50 -viewport 3d
Text output and log files can be opened in any text editor.
Clusterwise Correction for Multiple Comparisons
Note: The method used is based on: Smoothing and cluster thresholding for cortical surface-based group analysis of fMRI data. Hagler DJ Jr, Saygin AP, Sereno MI. NeuroImage (2006).
Remember to run the following commands in every new terminal window (aka shell) you open throughout this tutorial (see top of page for detailed instructions):
If You're at an Organized Course:
export SUBJECTS_DIR=$TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial cd $SUBJECTS_DIR/glm
If You're Not at an Organized Course:
<source_freesurfer> export TUTORIAL_DATA=<path_to_your_tutorial_data> export SUBJECTS_DIR=$TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial cd $SUBJECTS_DIR/glm
To perform a cluster-wise correction for multiple comparisons, we will run a simulation. The simulation is a way to get a measure of the distribution of the maximum cluster size under the null hypothesis. This is done by iterating over the following steps:
- Synthesize a z map.
- Smooth z map (using residual FWHM).
- Threshold z map (level and sign).
- Find clusters in thresholded map.
- Record area of maximum cluster.
Repeat over desired number of iterations (usually > 5,000).
In FreeSurfer, this information is stored in a simple text file called a CSD (Cluster Simulation Data) file.
Once we have the distribution of the maximum cluster size, we correct for multiple comparisons by:
- Going back to the original data.
- Thresholding using same level and sign.
- Finding clusters in thresholded map.
- For each cluster, p = probability of seeing a maximum cluster that size or larger during simulation.
Run the simulation
All these steps are performed with the command mri_glmfit-sim:
mri_glmfit-sim \ --glmdir lh.gender_age.glmdir \ --cache 4 neg \ --cwp 0.05\ --2spaces
Notes:
- Specify the same GLM directory (--glmdir).
- Use precomputed Z Monte Carlo simulation (--cache).
Vertex-wise/cluster-forming threshold of 4 (p < .0001).
- Specify the sign ("neg" for negative, "pos" for positive, or "abs" for absolute/unsigned).
--cwp 0.05 : Keep clusters that have cluster-wise p-values < 0.05. To see all clusters, set to .999.
- --2spaces : adjust p-values for two hemispheres (this assumes you will eventually look at the right hemisphere too).
- You can also generate your own precomputed data, but you only need to do this if you have a different average subject or you are not looking over all of the cortex.
- You can also use a permutation simulation instead of Z Monte Carlo. Run mri_glmfit-sim --help to get more information.
View the Corrected Results
In the contrast subdirectory, you will see several new files by running:
ls lh.gender_age.glmdir/lh-Avg-thickness-age-Cor
You will see the following new files:
cache.th40.neg.sig.pdf.dat -- probability distribution function of clusterwise correction cache.th40.neg.sig.cluster.mgh -- cluster-wise corrected map (overlay) cache.th40.neg.sig.cluster.summary -- summary of clusters (text) cache.th40.neg.sig.masked.mgh -- uncorrected sig values masked by the clusters that survive correction cache.th40.neg.sig.ocn.annot -- output cluster number (annotation of clusters) cache.th40.neg.sig.ocn.mgh -- output cluster number (segmentation showing where each numbered cluster is) cache.th40.neg.sig.voxel.mgh -- voxel-wise map corrected for multiple comparisons at a voxel (rather than cluster) level cache.th40.neg.sig.y.ocn.dat -- the average value of each subject in each cluster
First, look at the cluster summary (or click here):
less lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/cache.th40.neg.sig.cluster.summary
You can hit the 'Page Up' and 'Page Down' buttons or the 'Up' and 'Down' arrow keys to see the rest of the file. (To exit the less command, hit the 'q' button.)
Notes:
- This is a list of all the clusters that were found (20 of them).
- The CWP column is the cluster-wise probability (the number you are interested in). It is a simple p (ie, NOT -log10(p)).
- For example, cluster number 1 has a CWP of p=.0002.
Load the cluster annotation in freeview:
freeview -f $SUBJECTS_DIR/fsaverage/surf/lh.inflated:overlay=lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/cache.th40.neg.sig.cluster.mgh:overlay_threshold=2,5:annot=lh.gender_age.glmdir/lh-Avg-thickness-age-Cor/cache.th40.neg.sig.ocn.annot -viewport 3d
You should see clusters similar in shape to those pictured in the snapshots below. The color values associated with each cluster are arbitrary and may be different:
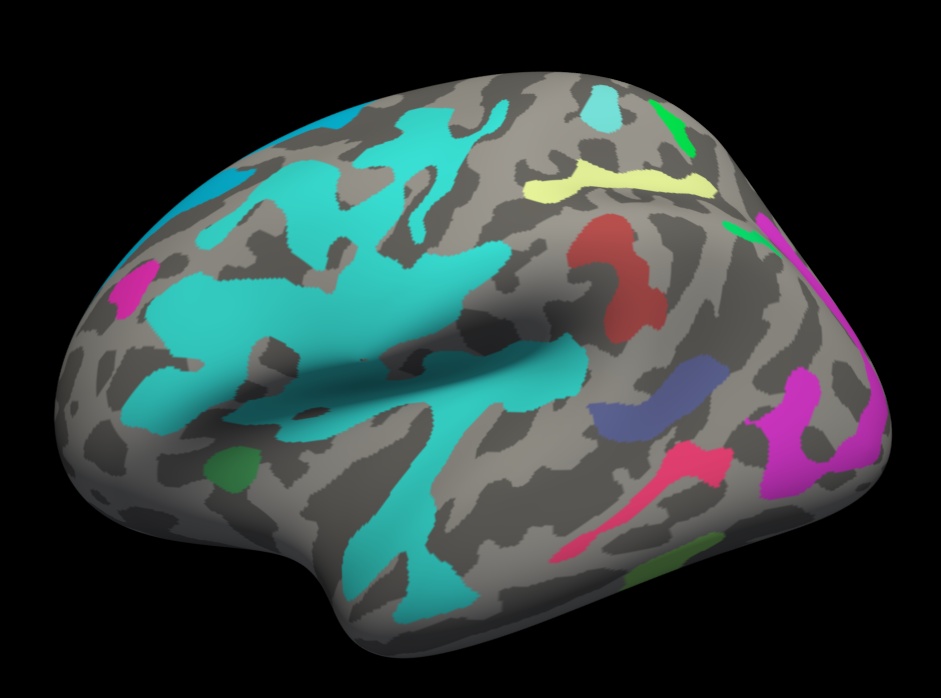
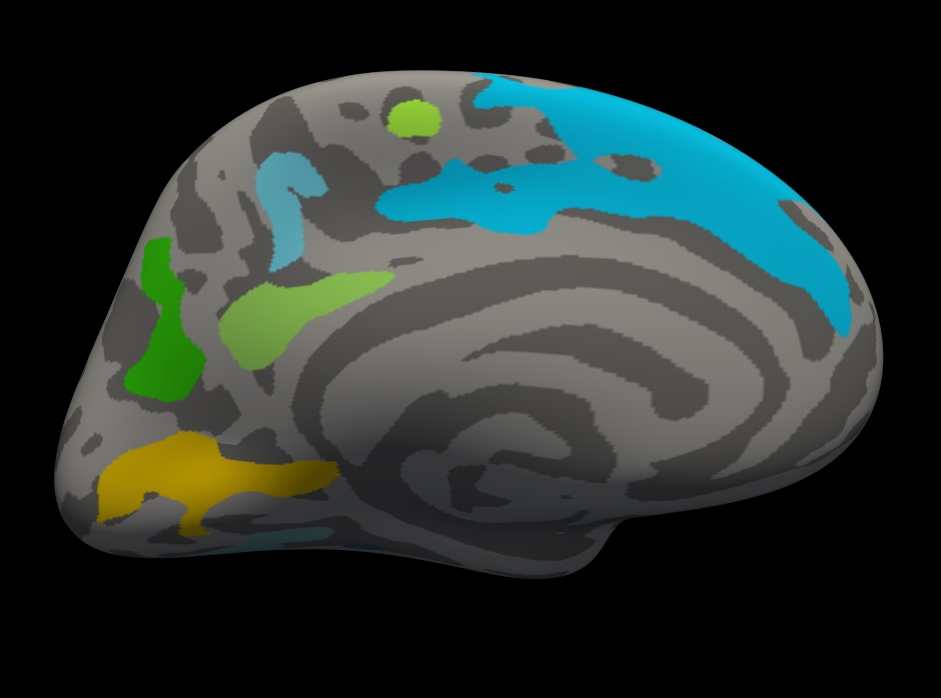
Notes:
- These are all clusters, regardless of significance.
- When you click on a cluster, the label will tell you the cluster number (eg, cluster-016).
Things to do:
- Find and click on cluster 1 (the largest cluster). It has a value of -3.69899 since this is log10(.0002). The -3.69899 is because the correlation is negative.
Find and click on cluster 17 (on the medial side of the brain). Its value is -1.4023 because this is log10(0.03960). Note that if you turn off the annotation, the cluster 17 is not visible because its significance is worse than the threshold we set (-fthresh 2, p < .01).
- All vertices within a cluster are the same value (the p-value of the cluster).
You can change the cluster-wise threshold by first clicking on "Show outline only" underneath the Annotation drop down menu. Then click on Configure (underneath "Overlay"), and set the Min value to your desired level. Alternatively, you can drag the red flag to adjust the cluster-wise threshold. As you do this, clusters will appear or disappear from the surface. (If your cursor is in the Min text box, the red flag won't move. Click on another text box to be able to move the flag.)
