|
Size: 5886
Comment:
|
Size: 7457
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 39: | Line 39: |
| Parameter estimates are also called "regression coefficients". | |
| Line 80: | Line 81: |
| == Assemble the Data == | == Assemble the Data (mris_preproc) == |
| Line 146: | Line 147: |
| == GLM Analysis == | == GLM Analysis (mri_glmfit) == |
| Line 169: | Line 170: |
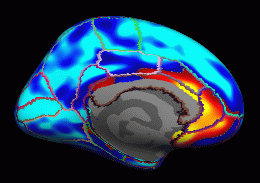
When this command is finished you will have an lh.gender_age.glmdir. There will be a number of output files in this directory, as well as two other directories. If you did an ''ls'' in your glmdir there will be: {{{ y.fsgd -- copy of input FSGD file (text) Xg.dat -- design matrix (text) mask.mgh -- binary mask (surface overlay) beta.mgh -- all parameter estimates (surface overlay) rstd.mgh -- residual standard deviation (surface overlay) sar1.mgh -- residual spatial AR1 (surface overlay) fwhm.dat -- average FWHM of residual (text) dof.dat -- degrees of freedom (text) lh-Avg-thickness-age-Cor -- contrast directory mri_glmfit.log -- log file (text, send this with bug reports) }}} There will be a subdirectory for each contrast that you specify. The name of the directory will be that of the contrast matrix file (without the .mtx extension). The '''lh-Avg-thickness-age-Cor''' directory will have the following files: {{{ C.dat -- orginal contrast matrix (text) gamma.mgh -- contrast effect size (surface overlay) F.mgh -- F ratio of contrast (surface overlay) sig.mgh -- significance, -log10(pvalue) (surface overlay) }}} {{{ tksurfer fsaverage lh inflated -annot aparc.annot \ -overlay lh.gender_age.glmdir/sig.mgh \ -fthresh 4 -fmid 5 }}} {{attachment:groupana.lat.uncor.th2.gif}} View->Configure->Overlay, set "Min" .01 {{attachment:groupana.med.uncor.th0.gif}} == Correction for Multiple Comparisons == |
Contents
Introduction
This tutorial is designed to introduce you to the "command-line" group analysis stream in FreeSurfer (as opposed to QDEC which is GUI-driven), including correction for multiple comparisons. While this tutorial shows you how to perform a surface-based thickness study, it is important to realize that most of the concepts learned here apply to any group analysis in FreeSurfer, surface or volume, thickness or fMRI.
QDEC Tutorial
FSGD Format
FSGD Examples
DODS vs DOSS
This Data Set
The data used for this tutorial is 40 subjects from Randy Buckner's lab. It consists of males and females ages 18 to 93. You can see the demograhics here. You will perform an analysis looking for the effect of age on cortical thickness accounting for the effects of gender in the analysis. This is the same data set used in the QDEC Tutorial, and you will get the same result.
General Linear Model (GLM) DODS Setup
Design Matrix/FSGD File
For this example, we will model the thickness as a straight line. A line has two parameters: intercept (or offset) and a slope.
- The slope is the change of thickness with age
- The intercept/offset is interpreted as the thickness at age=0.
Parameter estimates are also called "regression coefficients".
To account for effects of gender, we will model each sex with its own line, meaning that there will be four linear parameters (also called "betas"):
- Intercept for Females
- Intercept for Males
- Slope for Females
- Slope for Males
In FreeSurfer, this type of design is called DODS (for "Different-Offset, Different-Slope").
In FreeSurfer, you can either create your own design matrices, or, if you can specify your design in terms of a FreeSurfer Group Descriptor File (FSGD), FreeSurfer will create them for you. The FSGD file is a simple text file. See this page for the format. The [[FsTutorial/GroupAnalysisDemographics|demographics page]] also has an example FSGD file for this data.
Things to do:
- Create an FSGD file for the above design. One (gender_age.fsgd) already exists so that you can continue with the exercises.
Contrasts
A contrast is a vector that embodies the hypothesis we want to test. In this case, we wish to test the change in thickness with age after removing the effects of gender. To do this, create a simple text file with the following numbers:
0 0 0.5 0.5
Notes:
- There is one value for each parameter (so 4 values)
- The intercept/offset values are 0 (nuisance)
- The slope values are 0.5 so as to average the F and M slopes
- A file called lh-Avg-thickness-age-Cor.mtx already exists
Analysis
Assemble the Data (mris_preproc)
Assembling the data simply means:
- Resampling each subject's data into a common space, and
- Concatenating all the subjects' into a single file
- Spatial smoothing (can be done between 1 and 2)
This can be done in two equivalent ways:
Previously Cached (qcached) Data
When you have run recon-all with the -qcache option, recon-all will resample data onto the average subject (fsaverage) and smooth it at various FWHM (full-width/half-max), usually 0, 5, 10, 10, 20, and 25mm. This can speed later processing. The data for this tutorial have been cached, so run:
mris_preproc --fsgd gender_age.fsgd \ --cache-in thickness.fwhm10.fsaverage \ --target fsaverage --hemi lh \ --out lh.gender_age.thickness.10.mgh
Notes:
- Only takes a few seconds because the data have been cached
- The FSGD file lists all the subjects, helps keep order.
- The independent variable is the thickness smoothed to 10mm FWHM.
- The data are for the left hemisphere
- The output is lh.gender_age.thickness.10.mgh (which already exists)
Things to do:
- Run mri_info on lh.gender_age.thickness.10.mgh to find its dimensions.
Uncached Data
In the case that you have no cached the data, you can perform the same operations manually using the two commands below. There is no need to do this for this tutorial.
OPTIONAL: THIS WILL TAKE ABOUT 10 MINUTES mris_preproc --fsgd gender_age.fsgd \ --target fsaverage --hemi lh \ --meas thickness \ --out lh.gender_age.thickness.00.mgh
Notes:
- This resamples each subjects data to fsaverage
- Output is lh.gender_age.thickness.00.mgh, which is unsmoothed
OPTIONAL: THIS WILL TAKE ABOUT 5 MINUTES mri_surf2surf --hemi lh \ --s fsaverage \ --sval lh.gender_age.thickness.mgh \ --fwhm 10 \ --cortex \ --tval lh.gender_age.thickness.10B.mgh
- This smooths by 10mm FWHM
- "--cortex" means only smooth areas in cortex (exclude medial wall). This is automatically done with qcache. You can also specify other labels.
- Output is lh.gender_age.thickness.10B.mgh.
GLM Analysis (mri_glmfit)
mri_glmfit \ --y lh.gender_age.thickness.10.mgh \ --fsgd gender_age.fsgd dods\ --C lh-Avg-thickness-age-Cor.mtx \ --surf fsaverage lh \ --cortex \ --glmdir lh.gender_age.glmdir
Notes:
- Input is lh.gender_age.thickness.10.mgh
- Same FSGD used as with mris_preproc. Maintains subject order!
- DODS is specfied (it is the default)
- Only one contrast is used (lh-Avg-thickness-age-Cor.mtx) but you can specify multiple contrasts.
- "--cortex" specifies that the analysis only be done in cortex (ie, medial wall is zeroed out). Other labels can be used.
- The output directory is lh.gender_age.glmdir
- Should only take about 1min to run.
Things to do:
When this command is finished you will have an lh.gender_age.glmdir. There will be a number of output files in this directory, as well as two other directories. If you did an ls in your glmdir there will be:
y.fsgd -- copy of input FSGD file (text) Xg.dat -- design matrix (text) mask.mgh -- binary mask (surface overlay) beta.mgh -- all parameter estimates (surface overlay) rstd.mgh -- residual standard deviation (surface overlay) sar1.mgh -- residual spatial AR1 (surface overlay) fwhm.dat -- average FWHM of residual (text) dof.dat -- degrees of freedom (text) lh-Avg-thickness-age-Cor -- contrast directory mri_glmfit.log -- log file (text, send this with bug reports)
There will be a subdirectory for each contrast that you specify. The name of the directory will be that of the contrast matrix file (without the .mtx extension). The lh-Avg-thickness-age-Cor directory will have the following files:
C.dat -- orginal contrast matrix (text) gamma.mgh -- contrast effect size (surface overlay) F.mgh -- F ratio of contrast (surface overlay) sig.mgh -- significance, -log10(pvalue) (surface overlay)
tksurfer fsaverage lh inflated -annot aparc.annot \ -overlay lh.gender_age.glmdir/sig.mgh \ -fthresh 4 -fmid 5
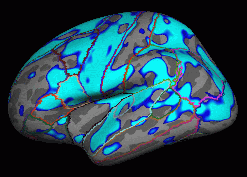
View->Configure->Overlay, set "Min" .01