| Deletions are marked like this. | Additions are marked like this. |
| Line 214: | Line 214: |
| Then, the software will write a (big!) bunch of files under the subject's mri directory, with the format: ''posterior_side_<substructure>_<T1>_<analysisID>_v2x.mgz''. Note that this posterior probabilities have nothing to do with anatomical "posterior". | Then, the software will write a (big!) bunch of files under the subject's mri directory, with the format: ''posterior_side_<substructure>_<T1>_<analysisID>_v2x.mgz''. Note that this posterior probabilities have nothing to do with anatomical '''posterior'''. |
Segmentation of hippocampal subfields and nuclei of the amygdala (cross-sectional and longitudinal)
This functionality has been available in the development version of FreeSurfer since August 31st 2017
Author: Juan Eugenio Iglesias
E-mail: iglesias [at] nmr.mgh.harvard.edu
Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu
If you use these tools in your analysis, please cite:
Hippocampus: A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. Iglesias, J.E., Augustinack, J.C., Nguyen, K., Player, C.M., Player, A., Wright, M., Roy, N., Frosch, M.P., Mc Kee, A.C., Wald, L.L., Fischl, B., and Van Leemput, K. Neuroimage, 115, July 2015, 117-137.
Amygdala: High-resolution magnetic resonance imaging reveals nuclei of the human amygdala: manual segmentation to automatic atlas. Saygin ZM & Kliemann D (joint 1st authors), Iglesias JE, van der Kouwe AJW, Boyd E, Reuter M, Stevens A, Van Leemput K, Mc Kee A, Frosch MP, Fischl B, Augustinack JC. Neuroimage, 155, July 2017, 370-382.
Longitudinal method: Bayesian longitudinal segmentation of hippocampal substructures in brain MRI using subject-specific atlases. Iglesias JE, Van Leemput K, Augustinack J, Insausti R, Fischl B, Reuter M. Neuroimage, 141, November 2016, 542-555.
See also: HippocampalSubfields, BrainstemSubstructures
Contents
- Motivation and General Description
- Installation
- Usage
- Gathering the volumes from all analyzed subjects
- Frequently asked questions
- Test data
1. Motivation and General Description
This tool is an evolution of our hippocampal module released with FreeSurfer 6.0. The tool uses a probabilistic atlas built with ultra-high resolution ex vivo MRI data (~0.1 mm isotropic) to produce an automated segmentation of the hippocampal substructures and the nuclei of the amygdala. The tool can use high-resolution images when available (typically, but not necessarily, T2 weighted). The main improvements with respect to the previous tool in FreeSurfer 6.0 are:
(a) Lower RAM memory requirements.
(b) In addition to the hippocampal subregions, it also simultaneously segments the nuclei of the amygdala (Saygin, Kliemann, et al., Neuroimage, 2017). Joint segmentation of hippocampus and amygdala ensures that structures do not overlap or leave gaps in between.
(c) It subdivides the hippocampal substructures into head and body, where applicable. For convenience, the code now generates an additional set of segmentations of each hemisphere at different levels of hierarchy (i.e., with merged labels). More specifically, it generates three additional sets of volumes with the following suffixes: HBT (head/body/tail), FS60, and CA. Segmentations without a hierarchy include all the (relevant) labels in the atlas: the 6.0 subfields divided in body/head where applicable, plus the amygdala (which is subdivided into lateral, basal, accessory basal, central, medial, cortical and paralaminar nuclei; and cortico-amngdaloid transition and anterior amygdala areas; see Saygin, Kliemann, et al., 2017 for details).
- HP Parcellation from Iglesias et al., 2015: the hippocampus is subdivided into head, body and tail.
HP Parcellation with additional head, body, tail: it mimics the FreeSurfer 6.0 hippocampal module, i.e., no head/body subdivision for the hippocampal subregions.
- Molecular subregion added to nearest neighbor: the "internal" labels (GC-ML-DG and molecular layer) are absorbed by the CA subfields. GC-ML-DG is absorbed by CA4, and the voxels in the molecular layer are assigned the label of the closest voxel that is neither molecular layer nor background
The way in which labels are merged in each of the three volumes is summarized in the following table:
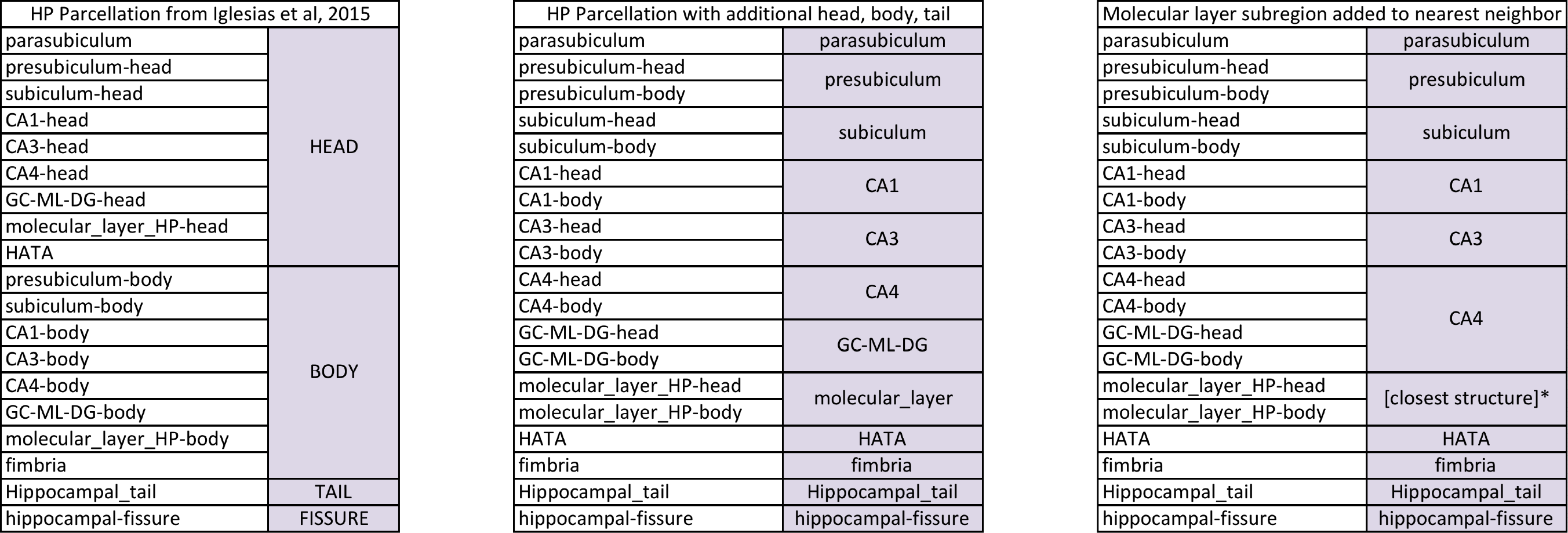
*: voxels in the molecular layer are assigned the label of the closest voxel that is neither molecular layer nor background
NOTE: CA2 IS ALWAYS INCLUDED IN CA3
In addition, the tool preserves all the advantages of its predecessor, including the accurate anatomical definitions derived from ultra-high resolution ex vivo scans, and the ability to adapt to the MRI contrast of the input data, even if multimodal.
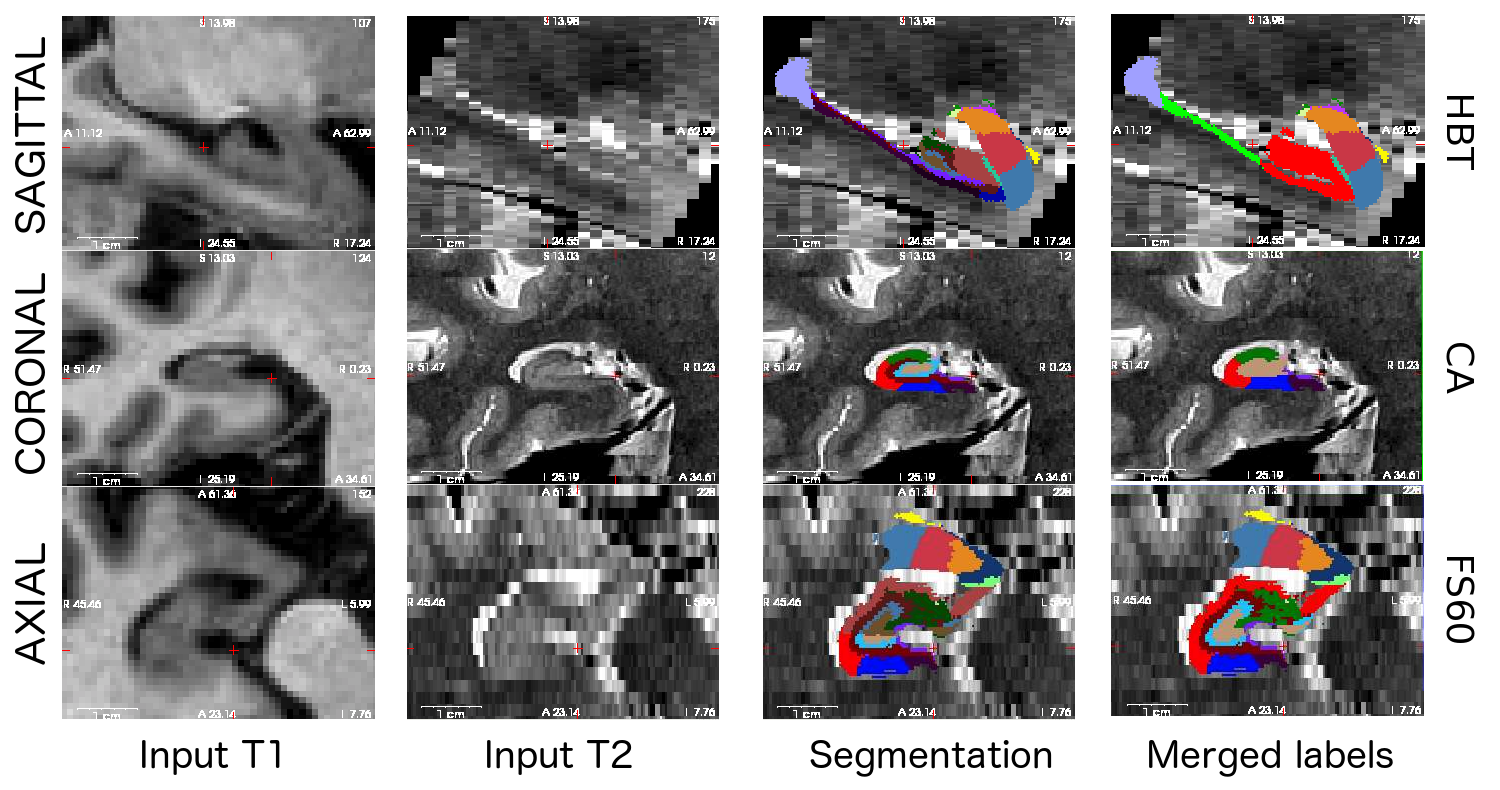
Here is a sample multimodal segmentation, computed from a 1mm (isotropic) T1 and a 0.4x0.4x2.0 mm (coronal) T2 scan, with the different hierarchical levels. Note that all nuclei of the amygdala are always shown (no merges).
2. Installation
The hippocampal module requires the Matlab R2012b (FreeSurfer 6.0 and dev versions until December 2017) or Matlab R2014b runtime (dev versions from January 2018 on); these runtimes are free, and therefore NO MATLAB LICENSES ARE REQUIRED TO USE THIS SOFTWARE. Please note that, if you have already installed the runtime for the brainstem module or the FS6.0 subfield module, you do not need to install it again. Instructions for the installation of the runtime can be found here:
https://surfer.nmr.mgh.harvard.edu/fswiki/MatlabRuntime
3. Usage
This software requires that a whole brain T1 scan of the subject has been analyzed with the main FreeSurfer stream ("recon-all"), i.e., we will assume that the command similar to this has already been run:
recon-all -all -s bert
where bert is the name of the subject. Bear in mind that, if this T1 scan has a voxel size smaller than 1 mm, it is possible to exploit this higher resolution by using the flag -cm in recon-all. Otherwise, both recon-all and this tool will work with a resampled version of the T1 volume at 1mm isotropic resolution. If we want to use the longitudinal version of this tool described in (Iglesias et al., Neuroimage, 2017), then it is also a requirement that the data have been processed with the longitudinal stream of FreeSurfer (LongitudinalProcessing).
This hippocampus/amygdala segmentation tool has three modes of operation: 1. Using only the main T1 volume processed with recon-all (norm.mgz); 2. Using an additional MRI volume (any MRI contrast supported) containing the hippocampus and amygdala (this scan is ideally of higher resolution); and 3. longitudinal segmentation of several time points of a single subject (only T1 mode supported at this time).
Using only the T1 scan from recon-all
To analyze your subject "bert", you would simply type:
segmentHA_T1.sh bert [SUBJECTS_DIR]
where the second argument is optional, and can be used to override the FreeSurfer subject directory.
The output will consist of the follwing set of files, which can be found under the subject's mri directory (in this example, $SUBJECTS_DIR/bert/mri/):
[lr]h.hippoSfVolumes-T1.v2x.txt: these text files store the estimated volumes of the hippocampal substructures and of the whole hippocampus.
[lr]h.amygNucVolumes-T1.v2x.txt: these text files store the estimated volumes of the nuclei of the amygdala and of the whole amygdala.
[lr]h.hippoAmygLabels-T1.v2x.mgz: they store the discrete segmentation volumes at subvoxel resolution (0.333 mm).
[lr]h.hippoAmygLabels-T1.v2x.FSvoxelSpace.mgz: they store the discrete segmentation volume in the FreeSurfer voxel space (normally 1mm isotropic, unless higher resolution data was used in recon-all with the flag -cm).
[lr]h.hippoAmygLabels-T1.v2x.[hierarchy].mgz: they store the segmentations with the different hierarchy levels.
[lr]h.hippoAmygLabels-T1.v2x.[hierarchy].FSvoxelSpace.mgz: same as above, but in FreeSurfer voxel space.
where [lr] refers to the left or right hemisphere, "v2x" is the version (v20 or, in newer dev versions, v21) and [hierarchy] refers to the different hierarchy levels in the segmentation, as described in Section 1 above ("HBT", "FS60 or "CA"). Discrete segmentation means a segmentation where there is one label for each voxel.
Note that [lr]h.hippoAmygLabels-T1.v2x.mgz and [lr]h.hippoAmygLabels-T1.v2x.[hierarchy].mgz cover only a patch around the hippocampus, at a higher resolution than the input image. The segmentation and the image are defined in the same physical coordinates, so you can visualize them simultaneously with (run from the subject's mri directory):
freeview -v nu.mgz -v lh.hippoAmygLabels-T1.v2x.mgz:colormap=lut -v rh.hippoAmygLabels-T1.v2x.mgz:colormap=lut freeview -v nu.mgz -v lh.hippoAmygLabels-T1.v2x.HBT.mgz:colormap=lut -v rh.hippoAmygLabels-T1.v2x.HBT.mgz:colormap=lut freeview -v nu.mgz -v lh.hippoAmygLabels-T1.v2x.FS60.mgz:colormap=lut -v rh.hippoAmygLabels-T1.v2x.FS60.mgz:colormap=lut freeview -v nu.mgz -v lh.hippoAmygLabels-T1.v2x.CA.mgz:colormap=lut -v rh.hippoAmygLabels-T1.v2x.CA.mgz:colormap=lut
On the other hand [lr]h.hippoAmygLabels-T1.v2x.FSvoxelSpace.mgz and [lr]h.hippoAmygLabels-T1.v2x.FSvoxelSpace.mgz live in the same voxel space as the other FreeSurfer volumes (e.g., orig.mgz, nu.mgz, aseg.mgz), so you can use it directly to produce masks for further analyses, but its resolution is lower (1 mm vs 0.333 mm).
Using an additional scan
If an additional MRI volume (e.g., a T2 scan, but it could be proton density, or even another T1) covering the hippocampi is available, we can use it to obtain a more reliable segmentation - particularly in the case in which its resolution is higher than that of the main T1 scan (even if anisotropic). In this case, the only requirement is that the additional scan is coarsely aligned to the main T1 scan.
The most important option of this mode of operation is whether the intensities of the main T1 volume (norm.mgz) should also be used in the segmentation, i.e., multispectral segmentation. Using the main T1 is advised when the additional scan is of comparable resolution as the T1, or when the additional scan does not cover the whole hippocampal region - in that case, the T1 information will be critical to segment the hippocampal regions outside the field of view of the additional volume (see for instance Figure 12e in the 2015 Neuroimage paper). Using the additional scan in isolation is advised when it is of higher resolution than the T1 while covering the whole hippocampi and amygdalae.
To analyze subject bert, the command would be:
segmentHA_T2.sh bert FILE_ADDITIONAL_SCAN ANALYSIS_ID USE_T1 [SUBJECTS_DIR]
FILE_ADDITIONAL_SCAN is the additional scan to use in the segmentation, in Nifty (.nii/.nii.gz) or FreeSurfer format (.mgh/.mgz). ANALAYSIS_ID is a user defined identifier that makes it possible to run different analysis with different types of additional scans. For example, you can run the command with a T2-weighted volume and use the identifier "T2", and then run it again with a PD-weighted volume and use the identifier "PD", such that both results will coexist in the subject's mri directory (see naming convention for the generated output below). For a certain modality of additional scan, make sure that you use the same ID for all the subjects within a study. Also please note that it is not possible to run two instances of segmentHA_T2.sh in parallel (one with USE_T1=0 and another with USE_T1=1) on the same subject with the same additional scan (segmentHA_T1.sh or segmentHA_T2 with a different additional scan are fine). USE_T1 is a flag that indicates whether the intensities of the main T1 scan should be used (multispectral segmentation). The words USE_T1 must be replaced with a 0 or 1 on the command line. SUBJECTS_DIR is optional, and overrides the FreeSurfer subject directory when provided.
These commands generate almost the same outputs as segmentHA_T1.sh in the subject's mri directory, with two differences:
(a) It also writes a volume with the additional scan rigidly aligned to the main T1 volume:
<analysisID>.FSspace.mgz: the additional scan, rigidly registered to the T1 data.
(b) The naming convention is:
[lr]h.hippoAmygLabels-<T1>-<analysisID>.v2x.[hierarchy].<FSvoxelSpace>.mgz
where <T1> appears only if USE_T1 = 1.
In order to visualize the outputs: let's say that you have run an analysis with id T2ADNI. Then, you can for example run the following command from the subject's mri directory:
freeview -v nu.mgz -v T2ADNI.FSspace.mgz:sample=cubic \ -v lh.hippoAmygLabels-T1-T2ADNI.v2x.mgz:colormap=lut -v rh.hippoAmygLabels-T1-T2ADNI.v2x.mgz:colormap=lut
* Longitudinal segmentation of T1 scans
Longitudinal analysis greatly reduces the confounding effect of inter-individual variability by using each subject as his or her own control. While one could analyze each time independently with segmentHA_T1.sh, such a cross-sectional analysis disregards the key fact that the scans are of the same subject. Instead, the scans corresponding to the different time points can be segmented jointly. Our method (Iglesias et al., Neuroimage, 2017) relies on a subject-specific atlas, and treats all time points the same way in order to avoid processing bias. We have shown in this publication that this strategy increases the robustness of the method and yields more sensitive subregion volumes. It is important to remark that, the same way as the main FreeSurfer longitudinal pipeline, this method does not assume any specific trajectory for the segmentations or corresponding volumes. Essentially, this means this method does not assume that the hippocampus continuously shrinks or grows, as some other methods do. This enables us to model e.g. cyclic behaviors. It is up to the user to incorporate such information in subsquent analyses, e.g., with a linear mixed effects model.
Let's say that <baseID> is the ID of the base subject (template) from the main stream. Then, we can produce the longitudinal hippocampal subregion segmentation with the following command:
segmentHA_T1_long.sh <baseID> [SUBJECTS_DIR]
where the second argument is optional, and can be used to override the FreeSurfer subject directory. The output of this script can be found under the corresponding mri directories of the longitudinally processed subjects (i.e., $SUBJECTS_DIR/<tpX>.long.<baseID>/mri/). The output files are very similar to those of segmentHA_T1.sh, with the difference that they will contain the suffix .long:
[lr]h.hippoAmygLabels-T1.long.v2x.[hierarchy].<FSvoxelSpace>.mgz: segmentations.
[lr]h.hippoSfVolumes-T1.long.v2x.txt: volumes of the hippocampal substructures.
[lr]h.amygNucVolumes-T1.long.v2x.txt: volumes of the nuclei of the amygdala
For visualization: bear in mind that the longitudinally processed time points are coregistered, so you can overlay their images and segmentations on top of each other. For instance, if you have two longitudinally processed subjects tp1.long.baseID and tp2.long.baseID, you could run (from SUBJECTS_DIR):
freeview -v tp1.long.baseID/mri/nu.mgz -v tp1.long.baseID/mri/lh.hippoAmygLabels-T1.long.v2x.mgz:colormap=lut -v tp1.long.baseID/mri/rh.hippoAmygLabels-T1.long.v2x.mgz:colormap=lut \
-v tp2.long.baseID/mri/nu.mgz -v tp2.long.baseID/mri/lh.hippoAmygLabels-T1.long.v2x.mgz:colormap=lut -v tp2.long.baseID/mri/rh.hippoAmygLabels-T1.long.v2x.mgz:colormap=lut
4. Gathering the volumes from all analyzed subjects
Once this module has been run on a number of subjects, it is possible to collect the volumes of the subregions of the hippocampus / amygdala of all subjects and write them to a single file - which can be easily read with a spreadsheet application. To do so, one would run:
quantifyHAsubregions.sh hippoSf <T1>-<analysisID> <output_file> <OPTIONAL_SUBJECTS_DIR> quantifyHAsubregions.sh amygNuc <T1>-<analysisID> <output_file> <OPTIONAL_SUBJECTS_DIR>
The first argument specifies whether we want to collect the volumes of the hippocampus (hippoSf) of the amygdala (amygNuc). The second argument is the name of the analysis: for the first mode of operation (only main T1 scans), it is simply T1. For the second mode (additional scan), it would be T1-<analysisID> (for multispectral analysis) or just <analysisID> (for segmentation based only on the additional scan). For longitudinal segmentation, it would just be T1.long. The argument <output_file> corresponds to the text file where the table with the volumes will be written. The fields are separated by spaces. Finally, the fourth argument is optional and overrides the FreeSurfer subjects directory.
5. Frequently asked questions (FAQ)
Does the longitudinal mode work with high-resolution T1s processed with the flat -cm?
Yes, but do not mix images of different resolutions (e.g., with and without -cm).
Are you sure that this software does not require Matlab licenses? Why does it require the Matlab runtime, then?
The software uses compiled Matlab code that requires the runtime (which is free), but no licenses. So, if you have a computer cluster, you can run hundred of segmentations in parallel without having to worry about Matlab licenses. And yes, this is all perfectly legal ![]()
The sum of the number of voxels of a given structure multiplied by the volume of a voxel is not equal to the volume reported in [lr]h.hippoSfVolumes*.txt.
This is because the volumes are computed upon a soft segmentation, rather than the discrete labels in [lr]h.hippoAmygLabels*.mgz. This is the same that happens with the main recon-all stream: if you compute volumes by counting voxels in aseg.mgz, you don't get the values reported in aseg.stats.
The size of the image volume of [lr]h.hippoAmygLabels*.mgz (in voxels) is not the same as that of norm.mgz or the additional input scan.
The segmentation [lr]h.hippoAmygLabels*.mgz covers only a patch around the hippocampus, at a higher resolution than the input image. The segmentation and the image are defined in the same physical coordinates, so that is why you can still visualize them simultaneously with FreeView using the commands listed above. The software also gives [lr]h.hippoAmygLabels*.FSvoxelSpace.mgz, which is in the same voxel space as the other FreeSurfer volumes, in case you need it to produce masks for other processing. But you are warned: these volumes are much uglier.
I am interested in the soft segmentations (i.e., posterior probabilities), can I have access to them?
Yes. All you need to do is to define an environment variable WRITE_POSTERIORS and set it to 1. For example, in tcsh or csh:
setenv WRITE_POSTERIORS 1
Or, in bash:
export WRITE_POSTERIORS=1
Then, the software will write a (big!) bunch of files under the subject's mri directory, with the format: posterior_side_<substructure>_<T1>_<analysisID>_v2x.mgz. Note that this posterior probabilities have nothing to do with anatomical posterior.
This module is CPU hungry
Indeed! The deformation of the atlas towards the input scan(s) is parallelized, and uses by default as many threads as available cores. If you want to limit the number of cores / threads that are used, you can do that through the environment variable ITK_GLOBAL_DEFAULT_NUMBER_OF_THREADS. For example, to use two cores (tcsh/csh):
setenv ITK_GLOBAL_DEFAULT_NUMBER_OF_THREADS 2
Or, in bash:
export ITK_GLOBAL_DEFAULT_NUMBER_OF_THREADS=2
What are the computational requirements to run this module?
If the input is just a standard resolution (1mm) T1 from the main FreeSurfer pipeline (i.e., no additional scans), then the software requires approximately 8GB of RAM memory to run. If a higher resolution T1 is processed with the flag -cm, or additional scans are used, then the exact amount of required RAM memory depends on the size and resolution of the scans. Likewise, in the longitudinal mode, the RAM requirements depend on the number of time points.
The volume of the whole hippocampus / whole amygdala obtained with this module is not equal to the value reported by the main FreeSurfer pipeline in $SUBJECTS_DIR/<subject_name>/stats/aseg.stats.
Yes! The reason for this is that the volumes correspond to two different analyses. We have found the estimates from this module to be slightly more accurate than FreeSurfer's in an Alzheimer's disease discrimination task (see paper).
What image formats are supported for the additional scan?
For now this software supports Nifti (.nii/.nii.gz) and FreeSurfer format (.mgh/.mgz).
Do you store the transform between the original additional volume and the FreeSurfer coordinate space?
Yes! You can find it under:
$SUBJECTS_DIR/<subject_name>/mri/transforms/T1_to_<analysisID>.v2x.lta
The registration between the T1 and the additional scan failed, i.e., the T1 and <analysisID>.FSspace.mgz do not overlap when I open them in Freeview
This can happen if the input additional scan and the T1 are not approximately registered in first place. Please align the additional scan to the T1 (without resampling, please!) and re-run the analysis. If you use Freeview (recommended), you can use tools -> Transform Volume to manually register the images, then hit the "save as" button and check the option "Do not resample voxel data when saving"
How can I quickly check whether the registration has failed?
In the subject's mri/transforms directory, the software writes an animated gif (T1_to_<analysisID>.v2x.QC.gif) that can be used as quality control. It flips back and forth between a central coronal slice in the T1 scan and its corresponding slice in the T2 scan. If the two images are not aligned, the problem can be fixed by prealigning the additional scan to the T1 scan, as explained above.
How reliable are the hippocampal substructure volumes estimated from standard resolution (1 mm) T1 data?
When segmenting 1 mm scans, the position of the internal boundaries between the hippocampal substructures largely relies on prior knowledge acquired from our ex vivo training data and summarized in our statistical atlas. For this reason, volumes of internal subregions (especially GC-DG, CA4 and molecular layer) must be interpreted with caution, and further validation with higher resolution data should ideally be carried out to confirm the results. On the other end of the spectrum, we have substructures such as the fimbria and tail, which are reliable at 1 mm.
How reliable are the volumes of the nuclei of the amygdala?
The position of the internal boundaries between the nuclei of the amygdala relies largely on prior knowledge acquired from our ex vivo training data and summarized in our statistical atlas. For this reason, volumes of the nuclei must be interpreted with caution.
Do the hippocampal subfields and amygdala nuclei need to be corrected for ICV (either by including ICV as a covariate or taking the ration if each subfield to the ICV)? If so, might it make more sense to correct them (perhaps take the ratio of each subfield to the hippocampus) for the volume of the hippocampus (after the hipp. itself has been corrected for the ICV)?
Yes, hippocampal subfields and amygdala nuclei need to be corrected for ICV. If you’re correcting the subfields by hippocampal volume: a) the question is a different one (“which fraction of the hippo volume is attributed to each subfield”); and b) I wouldn’t correct by ICV, because the values are already fractions / normalized (see point a).
6. Test data
Coming soon ...
