Bayesian Segmentation with Histological Atlas "NextBrain"
Visit the homepage of the NextBrain project for further information on this atlas.
Important: please download the latest development version of FreeSurfer to use this package
Author: Juan Eugenio Iglesias
E-mail: jiglesiasgonzalez [at] mgh.harvard.edu
Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu
Relevant publications:
"A probabilistic histological atlas of the human brain for MRI segmentation", Casamitjana et al., Nature, 2025. Paper on nature.com.
"Fast segmentation with the NextBrain histological atlas", Puonti et al., under review. Preprint available here.
Contents
- General Description
- Installation
- Basic usage
- Outputs
- Advanced options
- Frequently asked questions (FAQ)
1. General Description
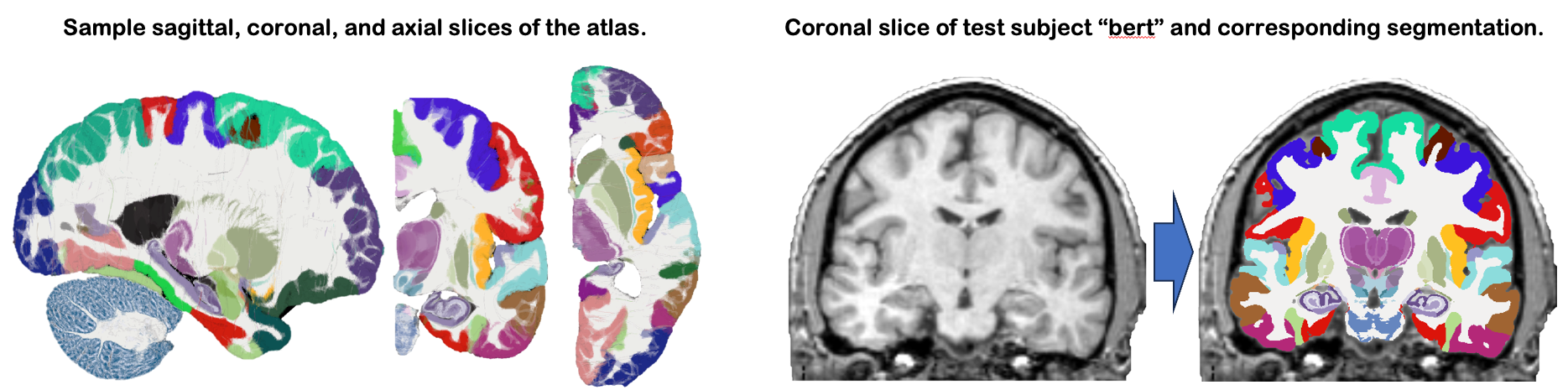
This module uses NextBrain, our new probabilistic atlas of the human brain, to segment ~300 distinct ROIs per hemisphere on in vivo or ex vivo scans (including single hemispheres). Segmentation relies on a Bayesian algorithm and is thus robust against changes in MRI pulse sequence (e.g., T1-weighted, T2-weighted, FLAIR, etc). Sample slices of the atlas and the segmentation of the sample subject "bert" are shown below:
2. Installation
The first time you run this module, it will prompt you to download the atlas. Follow the instructions on the screen to obtain the atlas files.
In addition: this module calls mri_super_synth; if you have never used this command before, it will also prompt you to download a model file.
3. Basic usage
The entry point of the module is the command mri_histo_atlas_segment_fireants, which implements the fast version of the algorithm. This version relies on FireANTs (Jena et al) for fast nonlinear registration of the atlas. The command line is:
mri_histo_atlas_segment_fireants --i INPUT_SCAN --o OUTPUT_DIRECTORY --device [cpu/cuda] --side [left/right] --mode [invivo/cerebrum/hemi/exvivo]
INPUT_SCAN: scan to process, in mgz or nii(.gz) format.
OUTPUT_DIRECTORY: directory where segmentations, volume files, etc, will be created (more on this below).
DEVICE: set to cpu or cuda.
SIDE: left or right. If you're analyzing both sides, you're better off running them sequentially (rather than in parallel) since the SuperSynth preprocessing will be reused when processing the second hemisphere.
MODE: type of scan: invivo, exvivo, cerebrum (ex vivo without brainstem or cerebellum), hemi (ex vivo with single cerebral hemisphere).
4. Outputs
The output directory will contain the following files:
seg.[left/right].nii.gz: segmentation of left/right hemisphere
lut.txt: the lookup table to visualize seg.[left/right].nii.gz, for convenience
vols.[left/right].csv: files with volumes of the brain regions segmented by the atlas, in CSV format.
SuperSynth: directory with segmentation of the scan at the whole structure level.
Additional flags: if advanced options are used (more details below).
5. Advanced options
The code also accepts the following optional flags:
- --bf_mode: Decides the bias field basis function model. Options: dct (default), polynomial, hybrid.
- --write_rgb: Save an RGB image based on the posterior probabilites to disk.
- --write_bias_corrected: Save the bias corrected input image to disk.
- --device_registration: Define a different device for the registration. Can be used to save GPU memory when working with an GPU with limited memory. Options: cpu, cuda. Default is the same as --device.
- --threads: Control the number of cpu thread used to run the algorithm. Default value is -1, which uses all available threads.
- --skip: An integer skipping (downsampling) factor for estimating the model parameters. More skipping saves memory, but sacrifices accuracy. Default: 1 (no skipping).
- --resolution: The resolution of the output segmentation. By default 0.4mm, which is higher than the typical input scan, to reduce aliasing.
- --smoothing_steps_HRmask: Number of smoothing steps used when upsampling the 1mm brain mask from BrainFM. More smoothing makes the outer border less jagged, but too much smoothing reduces accuracy. Default: 3.
- --skip_bf: Skip the bias field correction. Can be used to save memory if the input scan is already bias corrected or does not have a bias field (non MRI modality).
--smooth_grad_sigma: Gradient field smoothing parameter for the nonlinear FireAnts registration. Default: 1.0.
--smooth_warp_sigma: Warp field smoothing parameter for the nonlinear FireAnts registration. Default: 0.25.
--optimizer_lr: Learning rate for the nonlinear FireAnts registration optimizer. Default: 0.5.
- --cc_kernel_size: Size of the window for calculating the cross-correlation registration metric. Default: 7.
- --rel_weight_labeldiff: Relative weight for the Dice loss metric in the nonlinear registration. Default: 2.5.
- --save_atlas_nonlinear_reg: Save the nonlinearly registered atlas. Default: false.
- --save_field: Save the nonlinear deformation field. Default: false.
- --save_jacobian: Save the Jacobian determinant (log10) of the deformation field. Default: false.
- --yaml_path: path of custom YAML files to define groups of ROIs
Some notes:
* If you are running out of memory, using --skip 2 can help without sacrificing much accuracy. * The defaults --smooth_grad_sigma 1 and --smooth_warp_sigma 0.25 are pretty liberal and can cope with massive deformation, e.g., as in the Hip-CT images shown in the paper "Fast segmentation with the NextBrain". If you are working with a population without very strong atrophy or deformation, you can multiply those values by 2 in order to get more regular atlas deformation fields (you can explore the deformation with the --save_jacobian option).
Also: you can flexibly change the groupings of the modeled structures using the .yaml files under the /data_simplified folder. The structure groupings for the Gaussian Mixture modeled are controlled by two files: gmm_components_fireants.yaml and combined_atlas_labels_fireants.yaml. Let's say, as an example, that you wanted to add the internal segment of globus pallidus (label 206) as its own structure. To model it separately, you would first create a new class, called e.g., Internal Segment Pallidum, in the combined_atlas_labels_fireants.yaml file, and list label 206 under that structure (while removing it from the pallidum class). Next, you would add the class, with exactly the same name, to the gmm_components_fireants.yaml file and decide how many Gaussian distributions should be used to model its intensities. To make the non-linear registration aware of the contrast, you would add the structure, again with exactly the same name, to the file called recipe_intensities_cheating_image_fireants.yaml, and decide how its intensity should be generated from the seven structures than can be always reliably segmentation using BrainFM (see the file for examples).
6. Frequently asked questions (FAQ)
I have an ex vivo hemisphere with cerebellum and/or brainstem
If you use the hemi mode, you will not get the cerebellum or brainstem. Use the exvivo mode instead (with the caveat that you may lose some voxels around the medial wall, which may get assigned to the contralateral hemisphere).
Can the exvivo model handle arbitrary orientations of the input
No, it cannot. You need to manually reorient the brain to RAS (e.g., with Freeview).
Do I need a GPU?
Certainly not! The code should run in less than half an hour on any semi-modern workstation, if you allocate enough threads (or about two hours for an ex vivo scan at 0.25mm resolution).
What happened to the "full Bayesian" and "SynthMorph" versions?
To simplify the codebase, we are focusing on this method, which is fast but also versatile in terms of modeling / registration (as opposed to SynthMorph).
