Contents
Introduction
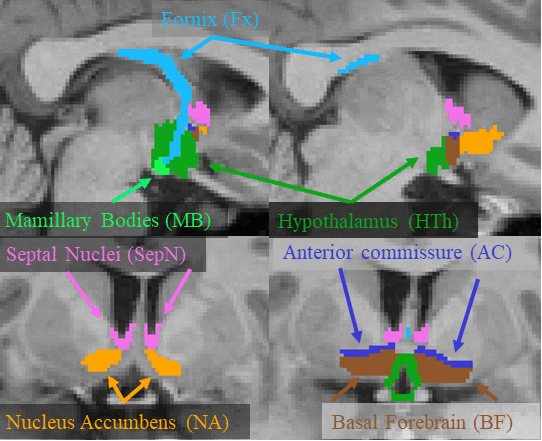
This page describes a deep learning tool, mri_sclimbic_seg, to automatically segment several subcorical limbic structures from T1-weighted images ("SC" = subcortical). The structures are shown in the figure below (note: anterior commissure is only present for reference reasons). Please cite: A Deep Learning Toolbox for Automatic Segmentation of Subcortical Limbic Structures from MRI Images Greve, DN, Billot, B, Cordero, D, Hoopes, M. Hoffmann, A, Dalca, A, Fischl, B, Iglesias, JE, Augustinack, JC. 2020. Submitted to Neuroimage.
Installation
Currently, you can obtain ScLimbic in two ways. One is to download the development version of FreeSurfer or you can download a "stripped-down" version (linux and mac) that has just what you need to run sclimbic, view results, and run group analysis. You do not need to run or understand FreeSurfer to run the basic ScLimbic, though ScLimbic can interface with FreeSurfer analysis. For the stripped-down version, you need to set it up with
cd /path-to-install tar xvfz sclimbic-linux-20210725.tar.gz export FREESURFER_HOME=/path-to-install/sclimbic source $FREESURFER_HOME/setup.sh
Basic Usage
In the basic usage, one passes the T1w volume (NIFTI or mgz format)one wants to segment and specifies the output:
mri_sclimbic_mri –i bert.mgz –o bert.sclimbic.mgz
This should take less than 1min to run. If the resolution is not 1mm3, then you must add the --conform. The segmentation can be viewed with freeview or tkmeditfv:
freeview bert.mgz bert.sclimbic.mgz
and should look like the figure above. Note that if you have the stripped-down version, you can find bert in $FREESURFER_HOME
Multiple Inputs
If one has multiple volumes, one can create a folder with the T1 volumes in (NIFTI or mgz format) one wants to segment, then run
mri_sclimbic_mri –i inputfolder –i outputfolder
The tool will find all the input images, segment them, and write out the segmentation volumes into the output folder. If the resolution is not 1mm3, then you must add the --conform flag. If the images are named subject1.mgz subject2.mgz, etc, the segmentations will be named subjec1.sclimbic.mgz, subject2.sclimbic.mgz, etc.
Table Output
Oftentimes, one wants to have the volume (in mm3) of the structures, eg, for use in a group analysis. To have ScLimbic produce a table for you, add --write_volumes to the command line: mri_sclimbic_mri –i inputfolder –i outputfolder --write_volumes
The above mri_sc_limbic command will also create a CSV file called sclimbic_volumes_all.csv where each row is a case, each column is a label, and each entry is the volume of that structure in mm3. each row is a case, each column is a label, and each entry is the volume of that structure in mm3.
Including Estimated Intracranial Volume (eTIV)
If one is going to perform a group analysis on the structure volume (eg, comparing subjects with Alzheimer's against healthy controls), then one should corrected for head size since the brain size scales with head size. If you do not have your own measure, you ScLimbic will compute one for you based on the method used in FreeSurfer based on Buckner, et al, 2004, NeuroImage 23:724-738. The simple way to do this is to just add --etiv to the mri_sclimbic_seg command line (note: this will increase the analysis time for each subject to about 5min -- but, hey, it's still pretty fast). When this is done, you will see an eTIV column in the CSV file. This will be automatically done when using ScLimbic in FreeSurfer mode.
Quality Control
It is important to check the quality of the segmentation as U-Nets can perform poorly when they see inputs that are very different than that they were trained on. The best way to QC the data is to simply visualize it using the freeview command above. However, this can be tedious when there are many subjects. Accordingly, ScLimbic can produce two QC files when run with --write_qa_stats:
mri_sclimbic_mri –i inputfolder –i outputfolder --write_volumes --write_qa_stats
This will produce sclimbic_zqa_scores_all.csv and sclimbic_confidence_all.csv The z-score should only be used for quality control. It should not be interpreted as a deviation from a normal population because the subjects used for manual labeling were not chosen with this in mind.
Group GLM Analysis
Running in FreeSurfer Mode
Other options
CPU threads, using the GPU, posteriors, AC
On a single threaded CPU, the program takes about 40sec to run on a single case; with 3 threads, the time drops to about 15sec; using a GPU does not reduce this significantly as much of the time is spent loading and writing. The tool uses about 20GB of memory. If the input volume is not 1mm3, then there is an option to reslice to this resolution; the reslicing is only internal – the output segmentation is resliced back to the original resolution. Note that this changing of resolution may affect the quality. If one is planning to perform a volumetric study, then one will need to normalize by ICV. If one does not have an estimate of the ICV, then the tool can compute it using the FreeSurfer method (Buckner et al., 2004); the ICV will be included as a column in the CSV file. Computing the ICV will increase the processing time to about 5min for each case. The CSV file can be imported into a statistical program like SPSS or R for further processing or it can be processed using FreeSurfer’s mri_glmfit, which includes automatic application of ICV correction if ICV is in the CSV. The user should visually spot check the segmentation output. To assist in quality control, the tool can output two more CSV files. One contains a z-score[1] for the volume each structure based on the means and standard deviations of the manual labels. In the other, the “confidence” (mean posterior probability within the label) is reported. If the z-score is very high or the confidence is very low, then the case should definitely be visually examined. The tool does not require knowledge of FreeSurfer; as long as FreeSurfer is installed properly, then the user need only understand and execute mri_sclimbic_seg.
[1] The z-score should only be used for quality control. It should not be interpreted as a deviation from a normal population because the subjects used for manual labeling were not chosen with this in mind.
