Joint segmentation of thalamic nuclei from T1 scan and DTI
PLEASE NOTE THAT:
1. AN UPDATED MODEL IS AVAILABLE FOR THE CNN WHICH HAS BEEN VALIDATED AGAINST ADDITIONAL DATASETS. IF YOU ARE USING FREESURFER 7.4, 7.4.1 OR A DEV VERSION COMPILED BEFORE 3RD JUN 2023 YOU MAY DOWNLOAD THE NEW MODEL HERE AND CALL IT WITH THE --model OPTION.
2. IMPROVEMENTS TO THE SEGMENTATION SCRIPT WERE IMPLEMENTED IN THE DEVELOPMENT BRANCH OF FREESURFER ON 2ND OCTOBER 2023. THIS VERSION OF THE TOOL MAY BE DOWNLOADED FROM THE GITHUB REPOSITORY. REPLACING THE RELEASE VERSION WITH THIS DOWNLOAD PREVENTS RARE OCCURRENCES OF FALSE POSITIVE THALAMIC SEGMENTATIONS. SEE FAQ FOR INSTRUCTIONS.
3. THESE PATCHES ARE LIVE IN THE NIGHTLY DEVELOPMENT BUILD OF FREESURFER AND WILL BE IN THE NEXT OFFICIAL RELEASE.
Authors: Henry Tregidgo and Juan Eugenio Iglesias
E-mail: h.tregidgo [at] ucl.ac.uk and jiglesiasgonzalez [at] mgh.harvard.edu
Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu
If you use these tools in your analysis, please cite:
* Accurate Bayesian segmentation of thalamic nuclei using diffusion MRI and an improved histological atlas. Tregidgo HFJ, Soskic S, Althonayan J, Maffei C, Van Leemput K, Golland P, Insausti R, Lerma-Usabiaga G, Caballero-Gaudes C, Paz-Alonso PM, Yendiki A, Alexander DC, Bocchetta M, Rohrer JD, Iglesias JE., NeuroImage, 274, 120129,(2023)
Additionally for the CNN implementation:
* Domain-agnostic segmentation of thalamic nuclei from joint structural and diffusion MRI. Tregidgo HFJ, Soskic S, Olchany MD, Althonayan J, Billot B, Maffei C, Golland P, Yendiki A, Alexander DC, Bocchetta M, Rohrer JD, Iglesias JE. In: Greenspan, H., et al. (eds.) MICCAI 2023. LNCS, vol 14227, pp. 247-257, Springer, Cham. (2023) (also on arXiv)
See also: ThalamicNuclei
Contents
- Motivation and General Description
- Bayesian tool
- Convolutional neural network (CNN)
- Frequently asked questions (FAQ)
1. Motivation and General Description
The ThalamicNuclei subfields module produces a parcellation of the thalamus into 25 different nuclei by applying a probabilistic atlas built with histological data to structural MRI volumes. However the lack of contrast in some structural scans can cause difficulty in correctly identifying boundaries that are more easily identifiable in modalities such as diffusion weighted MRI. For example, it often leaks on on to the corticospinal tract due to the lack of contrast on T1 images (while this is a boundary that is clearly visible in diffusion-weighted images):
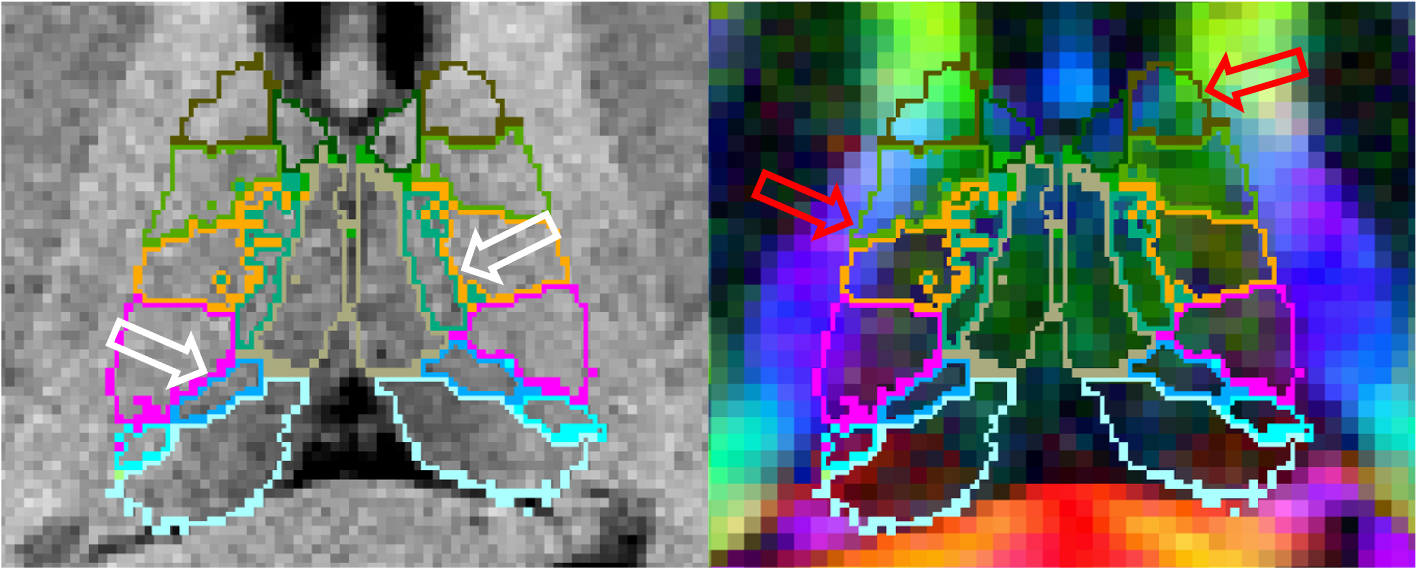
For this reason we have improved our method by incorporating information from diffusion imaging, specifically diffusion tensor imaging (DTI). To do this we not only incorporate a DTI likelihood term into our Bayesian framework but also modify our atlas to allow modelling of DTI specific inhomogeneities such as with white matter tracts or between the medial and lateral portions of the PuM. This results in improved identification of both interior and exterior thalamic boundaries.
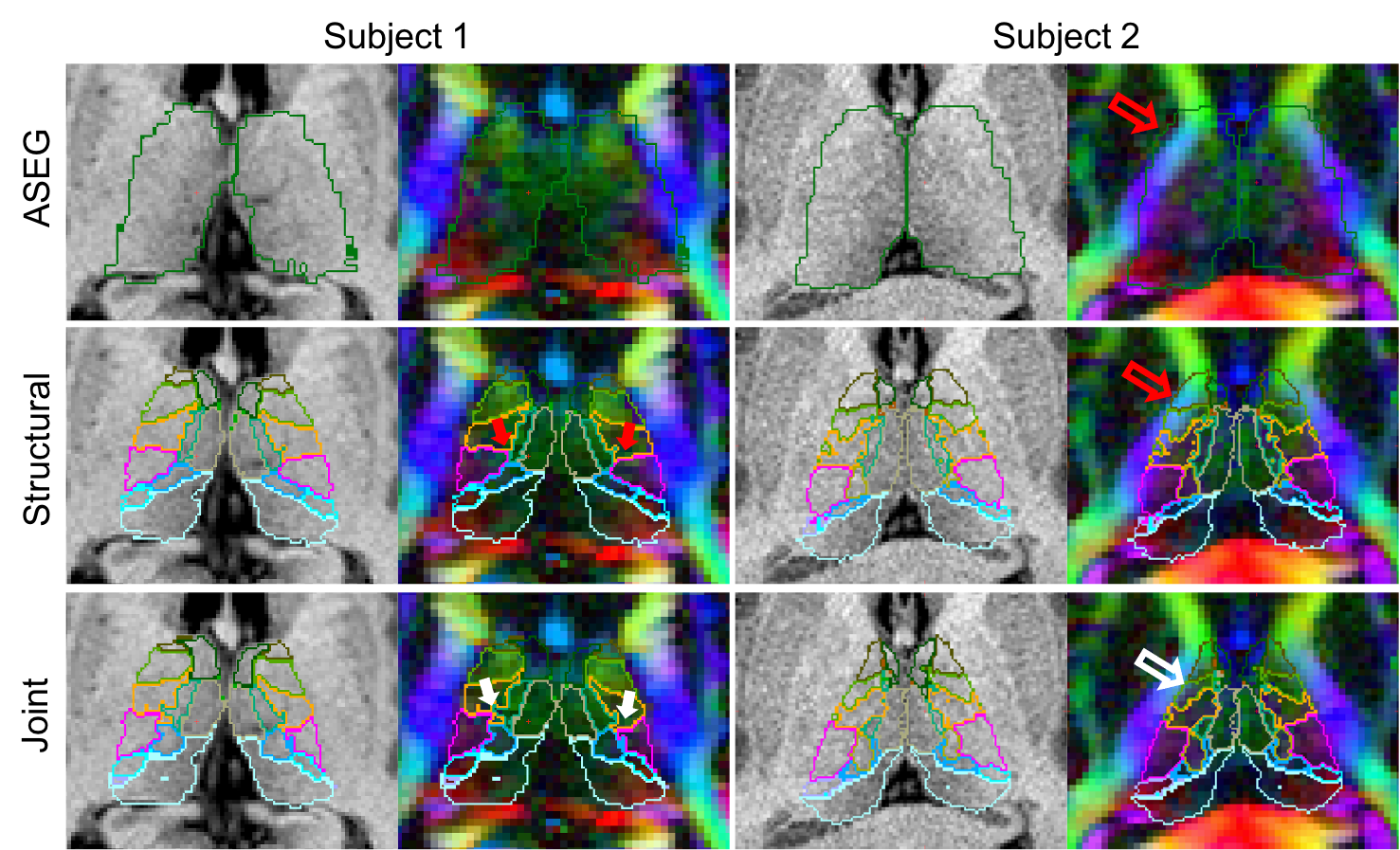
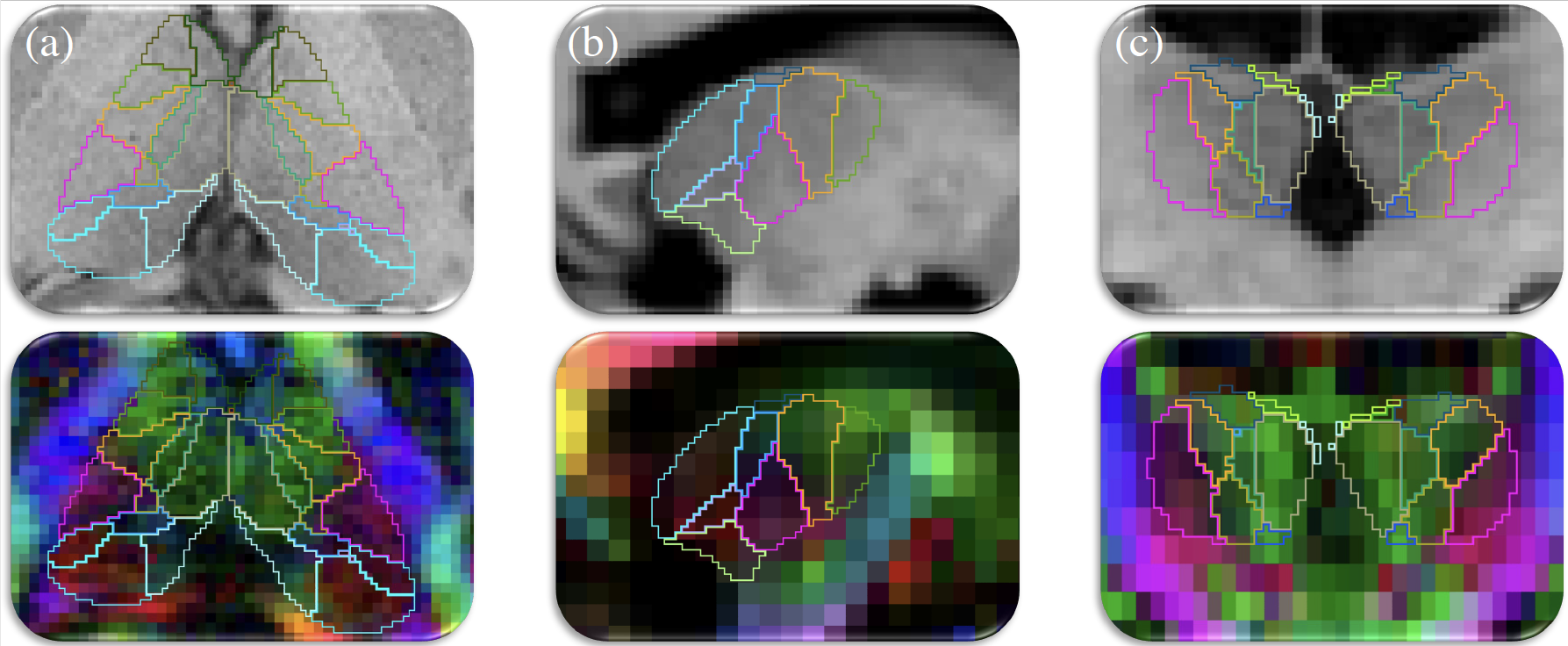
Additionally we address partial volume effects, present in DTI volumes with large voxels, by providing a convolutional neural network (CNN) that is trained to cope with large voxels of variable size. This CNN can segment the thalamic nuclei from T1 and diffusion MRI data obtained with virtually any acquisition, solving the problems posed by partial volume effects to Bayesian segmentations.
2. Bayesian Tool Usage (Development)
This module is currently being integrated into the Python framework that unifies the subregion segmentation modules and will be available soon. In the meantime, a compiled MATLAB binary will soon be introduced to the FreeSurfer development build and may be run using the free MATLAB runtime in the same fashion as the ThalamicNuclei module.
Following installation of the development build and MATLAB runtime, an example workflow would be:
Run FreeSurfer's main recon-all stream:
recon-all -all -s <subjid>
where <subjid> is the name of the subject.
Run Tracula:
trac-all -prep -s <subjid> -i <dicomfile>
where <dicomfile> is the path to the diffusion weighted MRI dicoms (see trac-all).
Call the bayesian joint segmentation shell script:
segmentThalamicNuclei_DTI.sh -s <subjid>
Options are also available to: specify DTI and output directories, prevent re-alignment of structural images from lta files, and specify that the DTI is of high resolution.
3. CNN Tool Usage
This software requires four registered input volumes, which are produced by the standard FreeSurfer/ TRACULA pipelines. These are:
A bias corrected whole brain T1 such as the norm.mgz output from the main FreeSurfer stream ("recon-all")
- A whole brain segmentation given by recon-all (aseg.mgz)
A fractional anisotropy volume such as dtifit_FA.nii.gz produced by the TRACULA module
A 4D volume containing the principle direction vector for each DTI voxel (eg. dtifit_V1.nii.gz from TRACULA)
An example workflow for obtaining these volumes would be:
Run FreeSurfer's main recon-all stream:
recon-all -all -s <subjid>
where <subjid> is the name of the subject.
Run Tracula:
trac-all -prep -s <subjid> -i <dicomfile>
where <dicomfile> is the path to the diffusion weighted MRI dicoms (see trac-all).
Apply the registrations to the structural images (note that we do not resample; we just update the header).
mri_vol2vol --mov <subjid>/mri/norm.mgz --o <subjid>/mri/norm.dwispace.mgz --lta <subjid>/dmri/xfms/anatorig2diff.bbr.lta --no-resample --targ <subjid>/dmri/dtifit_FA.nii.gz mri_vol2vol --mov <subjid>/mri/aseg.mgz --o <subjid>/mri/aseg.dwispace.mgz --lta <subjid>/dmri/xfms/anatorig2diff.bbr.lta --no-resample --targ <subjid>/dmri/dtifit_FA.nii.gz
Finally, run the CNN segmentation.
mri_segment_thalamic_nuclei_dti_cnn --t1 <subjid>/mri/norm.dwispace.mgz --aseg <subjid>/mri/aseg.dwispace.mgz --fa <subjid>/dmri/dtifit_FA.nii.gz --v1 <subjid>/dmri/dtifit_V1.nii.gz --o /path/to/outputSegmentation.nii.gz --vol /path/to/measuredVolumes.csv --threads <nthreads>
Where <nthreads> is the number of threads to use in processing.
The segmentations may then be viewed in Freeview using the following command
freeview <subjid>/mri/norm.dwispace.mgz path/to/outputSegmentation.nii.gz:colormap=LUT -dti <subjid>/dmri/dtifit_V1.nii.gz <subjid>/dmri/dtifit_FA.nii.gz
4. Frequently asked questions (FAQ)
How do I install the patched CNN script?
An updated version of the python script for CNN segmentation may be downloaded from the github repository. Once you have downloaded the new script you can apply the patch by replacing $FREESURFER_HOME/python/scripts/mri_segment_thalamic_nuclei_dti_cnn with the copy downloaded from github. Do not replace $FREESURFER_HOME/bin/mri_segment_thalamic_nuclei_dti_cnn
Why aren't the Pc, VM and Pt nuclei output by the CNN implementation?
These are the three smallest nuclei in our atlas, with volumes generally less than 2-3 mm^3. While they always have non-zero posterior probabilities in Bayesian segmentations, allowing us to estimate volume, they do not always "win" voxels in the hard segmentation. As such they don't appear in all the training examples for the CNN, so we exclude these labels to improve the robustness of the network.
Can I use DTI files generated outside of FreeSurfer?
Yes. Our tools expect the DTI to be provided in FSL format (ie. separate files for the FA, eigenvalues and principle directions of the tensors) and for the T1 weighted image to be aligned to the space of the DTI. The examples above show the filestructure locations of the relevant image and registration files generated by TRACULA. When using other software packages to generate these files, it is important to verify that they are provided in the correct format and that the images are correctly aligned.
Why might the segmentation not overlap with norm.mgz when I open them in Freeview?
Spatial re-alignment of the DTI requires re-orientation of the tensors, which can be non-trivial in the presence of any deformation (even affine). For this reason we work in the orientation of the DTI volumes while outputting at a higher resolution. For comparison to the T1 weighted input please view the re-aligned structural volume (eg. norm.dwispace.mgz) or apply the relevant spatial transform when loading into Freeview. (Note: please avoid re-sampling the T1 if possible, instead apply affine transformations to the volume headers.)
Can I substitute a SynthSeg segmentation for the ASEG to save time?
Your mileage may vary. While the segmentation provided by SynthSeg is compatible with our analysis, we have not incorporated T1 image bias into our model and so require the T1 source image to be bias corrected in a form similar to the norm.mgz from recon-all. We hope to further incorporate Synth tools into our analysis in a future update.
