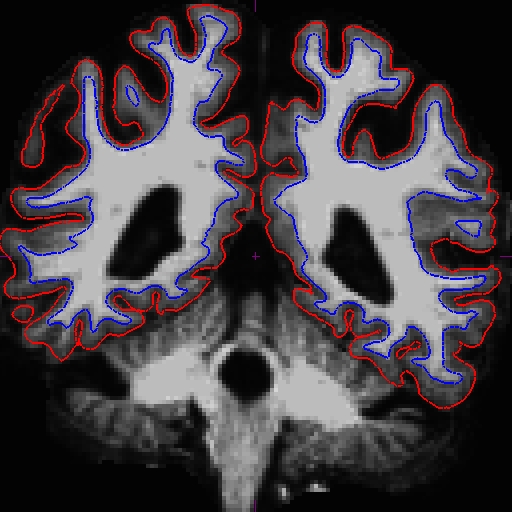
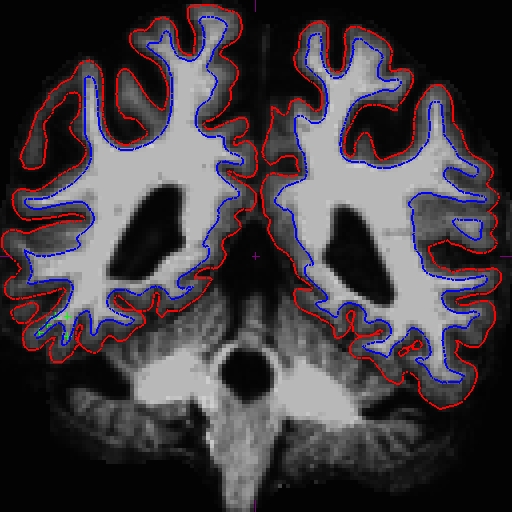
Using Control Points to Fix Intensity Normalization
To follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names.
Contents
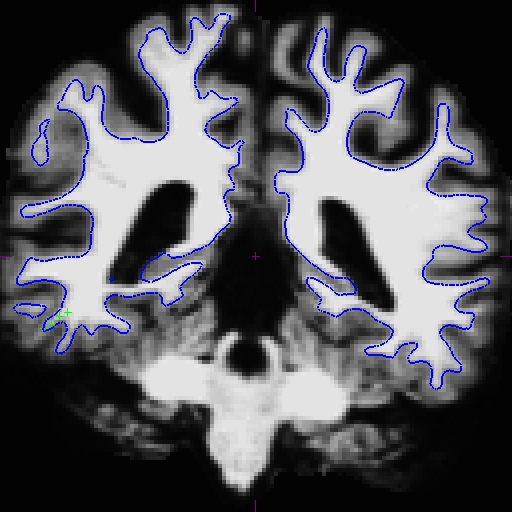
Notice that the intensity for the area in the bottom corner of the right hemisphere around slice 90 is only around 90 something instead of 110, and the surfaces here do not include all of the gm and wm. Because this problem occurs in both cross-sectionals, the base and subsequently the longitudinals are also affected. In cases like this, it depends on which data you are planning to use, you can choose to edit either the cross, base, or long. Control points work in all three stages.
In this example, we're demonstrating what the effect will be when adding control points at the very beginning of the stream - in the cross-sectionals. Find all the slices that have this problem and add control points carefully on the wm voxels.
Fixing the cross
Here is what we did. Left - OAS2_0121_MR1 (slice 87) and right - OAS2_0121_MR2 (slice 90).
Once you are satisfied with your control points placements, save it, and run the following commands to complete the fixed cross sectional runs:
Don't run these or any other recon-all commands on this page
It will take hours and has already been done for you.
recon-all -subjid OAS2_0121_MR1 -autorecon2 -autorecon3
recon-all -subjid OAS2_0121_MR2 -autorecon2 -autorecon3
After that is complete (or when the CA normalization step is complete - you can check in the recon-all-status.log), you can start rerunning the base (remember to delete or rename the base if it already existed, e.g. mv OAS2_0121 OAS2_0121_old). Or, if you prefer, you can wait until the recons are complete to check whether you are satisfied with the results before beginning the next step.
To inspect the fixed version of these two timepoints, run:
freeview -v OAS2_0121_MR1_fixed/mri/T1.mgz \
OAS2_0121_MR1_fixed/mri/brainmask.mgz \
-f OAS2_0121_MR1_fixed/surf/lh.pial:edgecolor=red \
OAS2_0121_MR1_fixed/surf/rh.pial:edgecolor=red \
OAS2_0121_MR1_fixed/surf/lh.white:edgecolor=blue \
OAS2_0121_MR1_fixed/surf/rh.white:edgecolor=bluefreeview -v OAS2_0121_MR2_fixed/mri/T1.mgz \
OAS2_0121_MR2_fixed/mri/brainmask.mgz \
-f OAS2_0121_MR2_fixed/surf/lh.pial:edgecolor=red \
OAS2_0121_MR2_fixed/surf/rh.pial:edgecolor=red \
OAS2_0121_MR2_fixed/surf/lh.white:edgecolor=blue \
OAS2_0121_MR2_fixed/surf/rh.white:edgecolor=blue
Fixing/Rerunning the base
To rerun the base, you would run this command:
recon-all -base OAS2_0121 -tp OAS2_0121_MR1 -tp OAS2_0121_MR2 -all
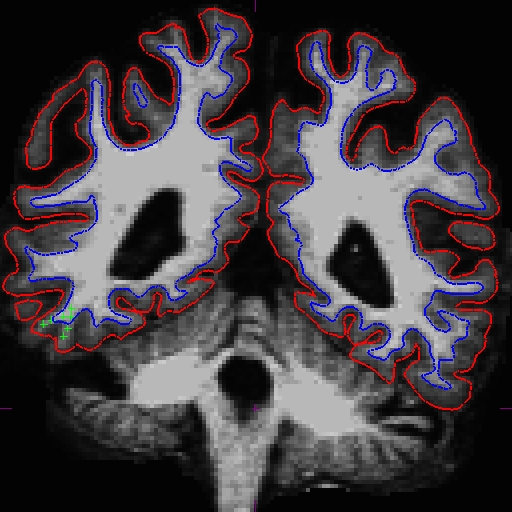
Once you are done with the base, check to see whether the surfaces are now including all white matter (we ran the above command for you, results are in *_intermediate):
freeview -v OAS2_0121_intermediate/mri/T1.mgz \
OAS2_0121_intermediate/mri/brainmask.mgz \
-f OAS2_0121_intermediate/surf/lh.pial:edgecolor=red \
OAS2_0121_intermediate/surf/rh.pial:edgecolor=red \
OAS2_0121_intermediate/surf/lh.white:edgecolor=blue \
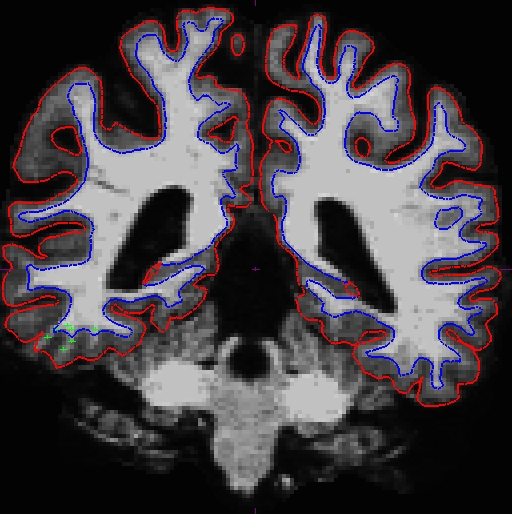
OAS2_0121_intermediate/surf/rh.white:edgecolor=blueNotice that although the surfaces improved, it is not perfect. In this case, you can add more control points to the base and would rerun with the command below (you would re-run the edited base without the _intermediate suffix as that is just a intermediate copy that we created for you):
recon-all -base OAS2_0121 -autorecon2 -autorecon3
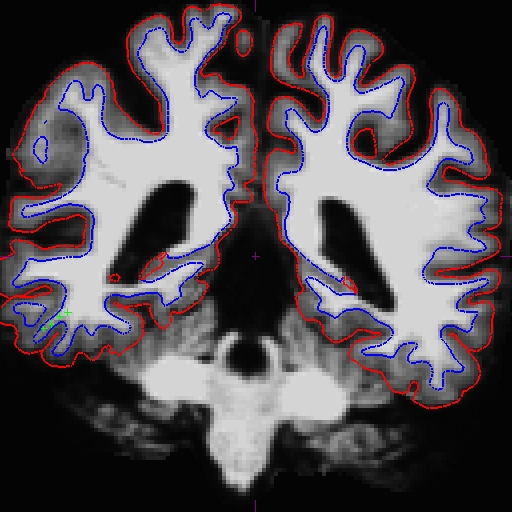
Now the base is much better. Compare these images at slice 109 (we removed the pial surface in order to have a better view). These are taken from OAS2_0121_intermediate (the mid point) and OAS2_0121_fixed (final results). You can open these using Freeview, and also the original base (OAS2_0121) if you want. You will not see the control points as shown on the intermediate base since this file is only the output and no interventions had taken place.
Rerunning the long
The last step is to rerun the longitudinals (also remove/rename existing longitudinal directories first), which would be done with these commands:
recon-all -long OAS2_0121_MR1 OAS2_0121 -all recon-all -long OAS2_0121_MR2 OAS2_0121 -all
Look at the output. We also re-ran the longs using the intermediate base from above to see what it would look like. You can open all three and compare using the following commands (you can do it for both MR1 and MR2):
freeview -v OAS2_0121_MR1.long.OAS2_0121/mri/T1.mgz \
OAS2_0121_MR1.long.OAS2_0121/mri/brainmask.mgz \
-f OAS2_0121_MR1.long.OAS2_0121/surf/lh.pial:edgecolor=red \
OAS2_0121_MR1.long.OAS2_0121/surf/rh.pial:edgecolor=red \
OAS2_0121_MR1.long.OAS2_0121/surf/lh.white:edgecolor=blue \
OAS2_0121_MR1.long.OAS2_0121/surf/rh.white:edgecolor=bluefreeview -v OAS2_0121_MR1.long.OAS2_0121_intermediate/mri/T1.mgz \
OAS2_0121_MR1.long.OAS2_0121_intermediate/mri/brainmask.mgz \
-f OAS2_0121_MR1.long.OAS2_0121_intermediate/surf/lh.pial:edgecolor=red \
OAS2_0121_MR1.long.OAS2_0121_intermediate/surf/rh.pial:edgecolor=red \
OAS2_0121_MR1.long.OAS2_0121_intermediate/surf/lh.white:edgecolor=blue \
OAS2_0121_MR1.long.OAS2_0121_intermediate/surf/rh.white:edgecolor=bluefreeview -v OAS2_0121_MR1.long.OAS2_0121_fixed/mri/T1.mgz \
OAS2_0121_MR1.long.OAS2_0121_fixed/mri/brainmask.mgz \
-f OAS2_0121_MR1.long.OAS2_0121_fixed/surf/lh.pial:edgecolor=red \
OAS2_0121_MR1.long.OAS2_0121_fixed/surf/rh.pial:edgecolor=red \
OAS2_0121_MR1.long.OAS2_0121_fixed/surf/lh.white:edgecolor=blue \
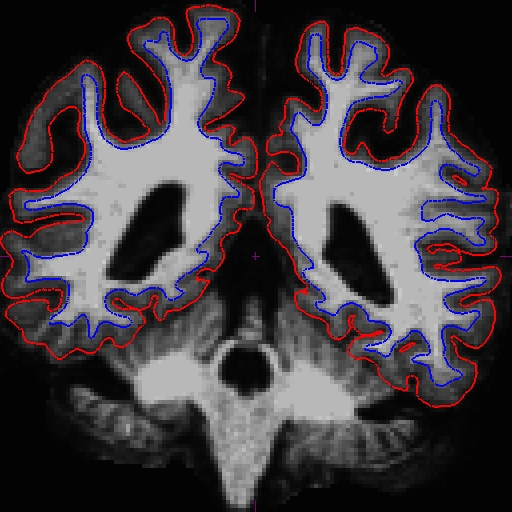
OAS2_0121_MR1.long.OAS2_0121_fixed/surf/rh.white:edgecolor=blueFor the longitudinals, control points were automatically copied over from the cross-sectionals, and the surfaces were initialized from the base, therefore making a great improvement on the surfaces at this end of the stream.
The before (left) and after (right) images from OAS2_0121_MR1.long.OAS2_0121 at slice 107.
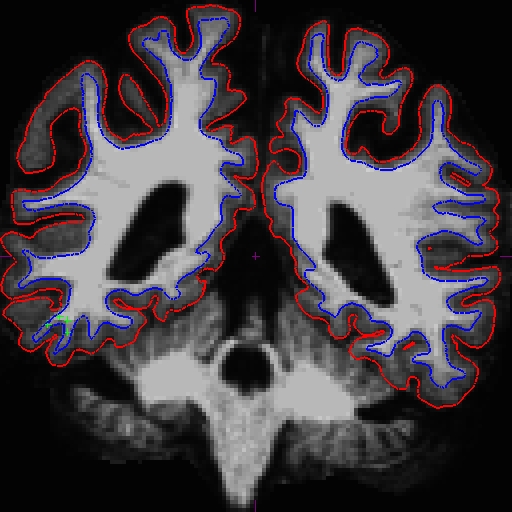
The before (left) and after (right) images from OAS2_0121_MR2.long.OAS2_0121 at slice 106.