Making FinalSurf edits
To follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names.
Here we will look at the second time point ( OAS2_0002_MR2). In the brain.finalsurfs.mgz volume (your aux volume in tkmedit) you can see the pial surface encroaching into the cerebellum in slices 65 - 68: Representative slice 68 shown below, problem at coordinates 111 136 68 in right hemisphere (and zoom-in):
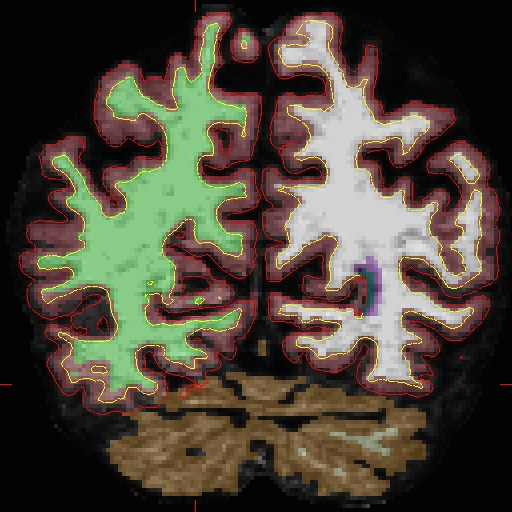
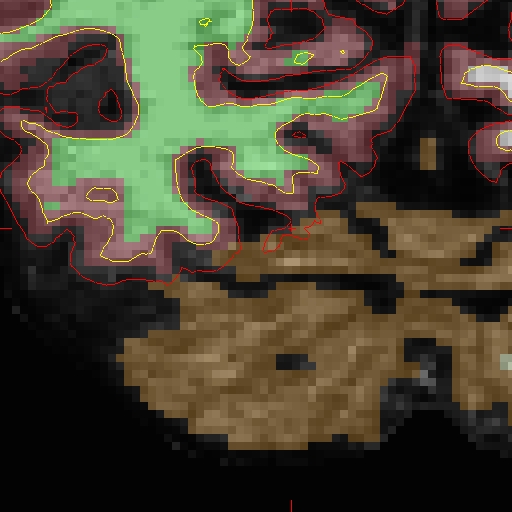
First make a copy of this volume to one called 'brain.finalsurfs.manedit.mgz' and save it within the mri directory of this time point, along with the other volumes.
cp OAS2_0002_MR2/mri/brain.finalsurfs.mgz OAS2_0002_MR2/mri/brain.finalsurfs.manedit.mgz
When recon-all is rerun, it will find the ...manedit.. file and knows that we want to incorporate manual edits. Next, load the brain.finalsurfs.manedit.mgz volume in Freeview (or as the aux volume in tkmedit) and edit this volume by going through the affected slices and removing the voxels in the cerebellum and surrounding areas that cause the pial surface misplacement. Once you are finished editing, save the changes to the edited volume. Next, run the following to have the edits take effect:
Don't run this recon-all command
It will take hours and has already been done for you.
recon-all -subjid OAS2_0002_MR2 -autorecon-pial
The results will look as follows, see OAS2_0002_MR2_fixed for the full data set:
freeview -v OAS2_0002_MR2_fixed/mri/brainmask.mgz \
OAS2_0002_MR2_fixed/mri/brain.finalsurfs.manedit.mgz \
OAS2_0002_MR2_fixed/mri/aseg.mgz:colormap=lut:opacity=0.25 \
-f OAS2_0002_MR2_fixed/surf/lh.pial:edgecolor=red \
OAS2_0002_MR2_fixed/surf/rh.pial:edgecolor=red \
OAS2_0002_MR2_fixed/surf/lh.white:edgecolor=blue \
OAS2_0002_MR2_fixed/surf/rh.white:edgecolor=blueNotice how the pial surface is now pulled in based on the deleted voxels (and zoom-in):
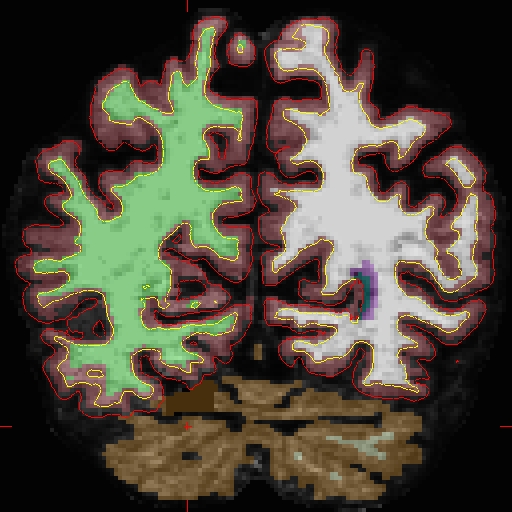
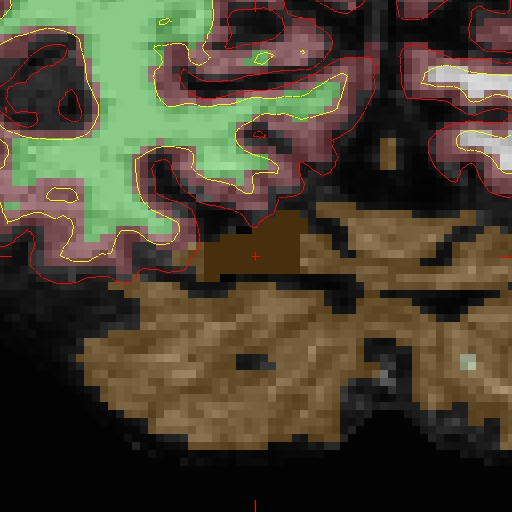
After looking at and fixing the problem in MR2, open MR1, and find the corresponding problem area. For OAS2_0002_MR1, make your copy of 'brain.finalsurfs.mgz' named 'brain.finalsurfs.manedit.mgz' in the mri directory with the following command:
cp OAS2_0002_MR1/mri/brain.finalsurfs.mgz OAS2_0002_MR1/mri/brain.finalsurfs.manedit.mgz
Look in a similar slice range as you had done for MR2:
freeview -v OAS2_0002_MR1/mri/brainmask.mgz \
OAS2_0002_MR1/mri/brain.finalsurfs.manedit.mgz \
OAS2_0002_MR1/mri/aseg.mgz:colormap=lut:opacity=0.25 \
-f OAS2_0002_MR1/surf/lh.pial:edgecolor=red \
OAS2_0002_MR1/surf/rh.pial:edgecolor=red \
OAS2_0002_MR1/surf/lh.white:edgecolor=blue \
OAS2_0002_MR1/surf/rh.white:edgecolor=blueYou can now fix the problem and compare your results to our fixed version:
freeview -v OAS2_0002_MR1_fixed/mri/brainmask.mgz \
OAS2_0002_MR1_fixed/mri/brain.finalsurfs.manedit.mgz \
OAS2_0002_MR1_fixed/mri/aseg.mgz:colormap=lut:opacity=0.25 \
-f OAS2_0002_MR1_fixed/surf/lh.pial:edgecolor=red \
OAS2_0002_MR1_fixed/surf/rh.pial:edgecolor=red \
OAS2_0002_MR1_fixed/surf/lh.white:edgecolor=blue \
OAS2_0002_MR1_fixed/surf/rh.white:edgecolor=blueNext, it is time to fix the base for this subject - return to the Longitudinal Tutorial to see how.
